Immune regulation by low doses of the DNA methyltransferase inhibitor 5-azacitidine in common human epithelial cancers
- PMID: 24583822
- PMCID: PMC3996658
- DOI: 10.18632/oncotarget.1782
Immune regulation by low doses of the DNA methyltransferase inhibitor 5-azacitidine in common human epithelial cancers
Abstract
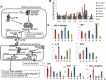
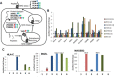
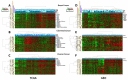
Epigenetic therapy is emerging as a potential therapy for solid tumors. To investigate its mechanism of action, we performed integrative expression and methylation analysis of 63 cancer cell lines (breast, colorectal, and ovarian) after treatment with the DNA methyltransferase inhibitor 5-azacitidine (AZA). Gene Set Enrichment Analysis demonstrated significant enrichment for immunomodulatory pathways in all three cancers (14.4-31.3%) including interferon signaling, antigen processing and presentation, and cytokines/chemokines. Strong upregulation of cancer testis antigens was also observed. An AZA IMmune gene set (AIMs) derived from the union of these immunomodulatory pathway genes classified primary tumors from all three types, into "high" and "low" AIM gene expression subsets in tumor expression data from both TCGA and GEO. Samples from selected patient biopsies showed upregulation of AIM genes after treatment with epigenetic therapy. These results point to a broad immune stimulatory role for DNA demethylating drugs in multiple cancers.
Trial registration: ClinicalTrials.gov NCT01105377 NCT01349959.
Figures


References
-
- Kaminskas E, Farrell A, Abraham S, Baird A, Hsieh LS, Lee SL, Leighton JK, Patel H, Rahman A, Sridhara R, Wang YC, Pazdur R. Approval summary: azacitidine for treatment of myelodysplastic syndrome subtypes. Clinical cancer research : an official journal of the American Association for Cancer Research. 2005;11(10):3604–3608. - PubMed
-
- Juergens RA, Wrangle J, Vendetti FP, Murphy SC, Zhao M, Coleman B, Sebree R, Rodgers K, Hooker CM, Franco N, Lee B, Tsai S, Delgado IE, Rudek MA, Belinsky SA, Herman JG, et al. Combination epigenetic therapy has efficacy in patients with refractory advanced non-small cell lung cancer. Cancer discovery. 2011;1(7):598–607. - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

