Quantitative proteomics analysis of signalosome dynamics in primary T cells identifies the surface receptor CD6 as a Lat adaptor-independent TCR signaling hub
- PMID: 24584089
- PMCID: PMC4037560
- DOI: 10.1038/ni.2843
Quantitative proteomics analysis of signalosome dynamics in primary T cells identifies the surface receptor CD6 as a Lat adaptor-independent TCR signaling hub
Abstract
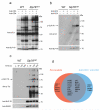
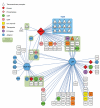
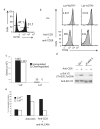
T cell antigen receptor (TCR)-mediated activation of T cells requires the interaction of dozens of proteins. Here we used quantitative mass spectrometry and activated primary CD4(+) T cells from mice in which a tag for affinity purification was knocked into several genes to determine the composition and dynamics of multiprotein complexes that formed around the kinase Zap70 and the adaptors Lat and SLP-76. Most of the 112 high-confidence time-resolved protein interactions we observed were previously unknown. The surface receptor CD6 was able to initiate its own signaling pathway by recruiting SLP-76 and the guanine nucleotide-exchange factor Vav1 regardless of the presence of Lat. Our findings provide a more complete model of TCR signaling in which CD6 constitutes a signaling hub that contributes to the diversification of TCR signaling.
Figures




Similar articles
-
Revisiting the Timing of Action of the PAG Adaptor Using Quantitative Proteomics Analysis of Primary T Cells.J Immunol. 2015 Dec 1;195(11):5472-81. doi: 10.4049/jimmunol.1501300. Epub 2015 Oct 28. J Immunol. 2015. PMID: 26512138
-
Mitogenic CD28 signals require the exchange factor Vav1 to enhance TCR signaling at the SLP-76-Vav-Itk signalosome.J Immunol. 2007 Feb 1;178(3):1363-71. doi: 10.4049/jimmunol.178.3.1363. J Immunol. 2007. PMID: 17237383
-
Age-dependent alterations in the assembly of signal transduction complexes at the site of T cell/APC interaction.J Immunol. 2000 Aug 1;165(3):1243-51. doi: 10.4049/jimmunol.165.3.1243. J Immunol. 2000. PMID: 10903722
-
Bridging the Gap: Modulatory Roles of the Grb2-Family Adaptor, Gads, in Cellular and Allergic Immune Responses.Front Immunol. 2019 Jul 25;10:1704. doi: 10.3389/fimmu.2019.01704. eCollection 2019. Front Immunol. 2019. PMID: 31402911 Free PMC article. Review.
-
Role of the LAT adaptor in T-cell development and Th2 differentiation.Adv Immunol. 2005;87:1-25. doi: 10.1016/S0065-2776(05)87001-4. Adv Immunol. 2005. PMID: 16102570 Review.
Cited by
-
Ubiquitylation as a Rheostat for TCR Signaling: From Targeted Approaches Toward Global Profiling.Front Immunol. 2015 Dec 16;6:618. doi: 10.3389/fimmu.2015.00618. eCollection 2015. Front Immunol. 2015. PMID: 26732666 Free PMC article. Review.
-
Mapping the SLP76 interactome in T cells lacking each of the GRB2-family adaptors reveals molecular plasticity of the TCR signaling pathway.Front Immunol. 2023 Mar 15;14:1139123. doi: 10.3389/fimmu.2023.1139123. eCollection 2023. Front Immunol. 2023. PMID: 37006259 Free PMC article.
-
The T cell CD6 receptor operates a multitask signalosome with opposite functions in T cell activation.J Exp Med. 2021 Feb 1;218(2):e20201011. doi: 10.1084/jem.20201011. J Exp Med. 2021. PMID: 33125054 Free PMC article.
-
Step-Wise Assembly of LAT Signaling Clusters Immediately After T Cell Receptor Triggering Contributes to Signal Propagation.Int J Mol Sci. 2025 Apr 25;26(9):4076. doi: 10.3390/ijms26094076. Int J Mol Sci. 2025. PMID: 40362315 Free PMC article.
-
Alternative ZAP70-p38 signals prime a classical p38 pathway through LAT and SOS to support regulatory T cell differentiation.Sci Signal. 2019 Jul 23;12(591):eaao0736. doi: 10.1126/scisignal.aao0736. Sci Signal. 2019. PMID: 31337738 Free PMC article.
References
-
- Roncagalli R, Mingueneau M, Gregoire C, Malissen M, Malissen B. LAT signaling pathology: an “autoimmune” condition without T cell self-reactivity. Trends Immunol. 2010;31:253–259. - PubMed
-
- Sommers CL, et al. A LAT mutation that inhibits T cell development yet induces lymphoproliferation. Science. 2002;296:2040–2043. - PubMed
-
- Mingueneau M, et al. Loss of the LAT adaptor converts antigen-responsive T cells into pathogenic effectors that function independently of the T cell receptor. Immunity. 2009;31:197–208. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

