The transcription factor BATF operates as an essential differentiation checkpoint in early effector CD8+ T cells
- PMID: 24584090
- PMCID: PMC4000237
- DOI: 10.1038/ni.2834
The transcription factor BATF operates as an essential differentiation checkpoint in early effector CD8+ T cells
Abstract
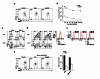
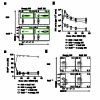
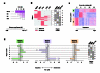
The transcription factor BATF is required for the differentiation of interleukin 17 (IL-17)-producing helper T cells (TH17 cells) and follicular helper T cells (TFH cells). Here we identified a fundamental role for BATF in regulating the differentiation of effector of CD8(+) T cells. BATF-deficient CD8(+) T cells showed profound defects in effector population expansion and underwent proliferative and metabolic catastrophe early after encountering antigen. BATF, together with the transcription factors IRF4 and Jun proteins, bound to and promoted early expression of genes encoding lineage-specific transcription-factors (T-bet and Blimp-1) and cytokine receptors while paradoxically repressing genes encoding effector molecules (IFN-γ and granzyme B). Thus, BATF amplifies T cell antigen receptor (TCR)-dependent expression of transcription factors and augments the propagation of inflammatory signals but restrains the expression of genes encoding effector molecules. This checkpoint prevents irreversible commitment to an effector fate until a critical threshold of downstream transcriptional activity has been achieved.
Figures
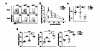



Similar articles
-
A Critical Role of IL-21-Induced BATF in Sustaining CD8-T-Cell-Mediated Chronic Viral Control.Cell Rep. 2015 Nov 10;13(6):1118-1124. doi: 10.1016/j.celrep.2015.09.069. Epub 2015 Oct 29. Cell Rep. 2015. PMID: 26527008 Free PMC article.
-
BATF-JUN is critical for IRF4-mediated transcription in T cells.Nature. 2012 Oct 25;490(7421):543-6. doi: 10.1038/nature11530. Epub 2012 Sep 19. Nature. 2012. PMID: 22992523 Free PMC article.
-
The activating protein 1 transcription factor basic leucine zipper transcription factor, ATF-like (BATF), regulates lymphocyte- and mast cell-driven immune responses in the setting of allergic asthma.J Allergy Clin Immunol. 2014 Jan;133(1):198-206.e1-9. doi: 10.1016/j.jaci.2013.09.049. Epub 2013 Nov 28. J Allergy Clin Immunol. 2014. PMID: 24290279
-
The transcription factor BATF modulates cytokine-mediated responses in T cells.Cytokine Growth Factor Rev. 2016 Aug;30:39-45. doi: 10.1016/j.cytogfr.2016.03.004. Epub 2016 Mar 6. Cytokine Growth Factor Rev. 2016. PMID: 26970726 Review.
-
Genes associated with T helper 17 cell differentiation and function.Front Biosci (Elite Ed). 2016 Jun 1;8(3):427-35. doi: 10.2741/E777. Front Biosci (Elite Ed). 2016. PMID: 27100349 Review.
Cited by
-
Regulatory circuits of T cell function in cancer.Nat Rev Immunol. 2016 Oct;16(10):599-611. doi: 10.1038/nri.2016.80. Epub 2016 Aug 16. Nat Rev Immunol. 2016. PMID: 27526640 Review.
-
TCF-1-Centered Transcriptional Network Drives an Effector versus Exhausted CD8 T Cell-Fate Decision.Immunity. 2019 Nov 19;51(5):840-855.e5. doi: 10.1016/j.immuni.2019.09.013. Epub 2019 Oct 9. Immunity. 2019. PMID: 31606264 Free PMC article.
-
Transcriptional Modulation of the Host Immunity Mediated by Cytokines and Transcriptional Factors in Plasmodium falciparum-Infected Patients of North-East India.Biomolecules. 2019 Oct 11;9(10):600. doi: 10.3390/biom9100600. Biomolecules. 2019. PMID: 31614626 Free PMC article.
-
A data-driven Boolean model explains memory subsets and evolution in CD8+ T cell exhaustion.NPJ Syst Biol Appl. 2023 Jul 31;9(1):36. doi: 10.1038/s41540-023-00297-2. NPJ Syst Biol Appl. 2023. PMID: 37524735 Free PMC article.
-
BATF represses BIM to sustain tolerant T cells in the periphery.J Exp Med. 2023 Dec 4;220(12):e20230183. doi: 10.1084/jem.20230183. Epub 2023 Oct 20. J Exp Med. 2023. PMID: 37862030 Free PMC article.
References
-
- Kaech SM, Wherry EJ, Ahmed R. Effector and memory T-cell differentiation: implications for vaccine development. Nat Rev Immunol. 2002;2(4):251–262. - PubMed
-
- Rutishauser RL, Kaech SM. Generating diversity: transcriptional regulation of effector and memory CD8 T-cell differentiation. Immunol Rev. 2010;235(1):219–233. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

