Genetic diversity analysis reveals that geographical environment plays a more important role than rice cultivar in Villosiclava virens population selection
- PMID: 24584249
- PMCID: PMC3993286
- DOI: 10.1128/AEM.03936-13
Genetic diversity analysis reveals that geographical environment plays a more important role than rice cultivar in Villosiclava virens population selection
Abstract
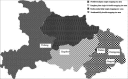
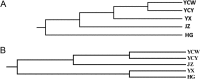
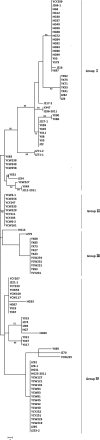
Rice false smut caused by Villosiclava virens is an economically important disease of grains worldwide. The genetic diversity of 153 isolates from six fields located in Wuhan (WH), Yichang Wangjia (YCW), Yichang Yaohe (YCY), Huanggang (HG), Yangxin (YX), and Jingzhou (JZ) in Hubei province of China were phylogenetically analyzed to evaluate the influence of environments and rice cultivars on the V. virens populations. Isolates (43) from Wuhan were from two rice cultivars, Wanxian 98 and Huajing 952, while most of the other isolates from fields YCW, YCY, HG, YX, and JZ originated from different rice cultivars with different genetic backgrounds. Genetic diversity of isolates was analyzed using random amplified polymorphic DNA (RAPD) and single-nucleotide polymorphisms (SNP). The isolates from the same cultivars in Wuhan tended to group together, indicating that the cultivars had an important impact on the fungal population. The 110 isolates from individual fields tended to cluster according to geographical origin. The values of Nei's gene diversity (H) and Shannon's information index (I) showed that the genetic diversity among isolates was higher between than within geographical populations. Furthermore, mean genetic distance between groups (0.006) was higher than mean genetic distance within groups (0.0048) according to MEGA 5.2. The pairwise population fixation index (FST) values also showed significant genetic differentiation between most populations. Higher genetic similarity of isolates from individual fields but different rice cultivars suggested that the geographical factor played a more important role in the selection of V. virens isolates than rice cultivars. This information could be used to improve the management strategy for rice false smut by adjusting the cultivation measures, such as controlling fertilizer, water, and planting density, in the rice field to change the microenvironment.
Figures



References
-
- White JF, Jr, Sullivan R, Moy M, Patel R, Duncan R. 2000. An overview of problems in the classification of plant-parasitic Clavicipitaceae. Stud. Mycol. 45:95–105 http://www.fungalbiodiversitycentre.com/publications/1045/content/pdf/09....
-
- Tanaka E, Ashizawa T, Sonoda R, Tanaka C. 2008. Villosiclava virens gen. nov., comb. nov., teleomorph of Ustilaginoidea virens, the causal agent of rice false smut. Mycotaxon 106:491–501 http://www.researchgate.net/publication/234130822
-
- Ashizawa T, Takahashi M, Arai M, Arie T. 2012. Rice false smut pathogen, Ustilaginoidea virens, invades through small gap at the apex of a rice spikelet before heading. J. Gen. Plant Pathol. 78:255–259. 10.1007/s10327-012-0389-3 - DOI
-
- Tang YX, Jin J, Hu DW, Yong ML, Xu Y, He LP. 2013. Elucidation of the infection process of Ustilaginoidea virens (teleomorph: Villosiclava virens) in rice spikelets. Plant Pathol. 62:1–8. 10.1111/j.1365-3059.2012.02629.x - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

