Inhibition of post-translational N-glycosylation by HRD1 that controls the fate of ABCG5/8 transporter
- PMID: 24584735
- PMCID: PMC3939451
- DOI: 10.1038/srep04258
Inhibition of post-translational N-glycosylation by HRD1 that controls the fate of ABCG5/8 transporter
Abstract
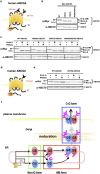
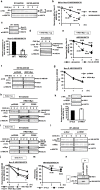
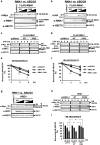
N-glycosylation of proteins in endoplasmic reticulum is critical for protein quality control. We showed here a post-translational N-glycosylation affected by the HRD1 E3 ubiquitin ligase. Both WT- and E3-defective C329S-HRD1 decreased the level of high mannose form of ABCG8, a protein that heterodimerizes with ABCG5 to control sterol balance. Meanwhile, HRD1 increased the non-glycosylated ABCG8 regardless of its E3 activity, thereby suppressing full maturation of ABCG5/8 transporter. Pulse chase and mutational analysis indicated that HRD1 inhibits STT3B-dependent post-translational N-glycosylation of ABCG8. Whereas, HRD1 had only slight effect on the N-glycosylation status of ABCG5; rather it accelerated ABCG5 degradation in an E3 activity-dependent manner. Finally, RMA1, another E3 ubiquitin ligase, accelerated the degradation of both ABCG5 and ABCG8 via E3 activity-dependent manner. HRD1 and RMA1 may therefore be negative regulators of disease-associated transporter ABCG5/ABCG8. The findings also highlight the unexpected E3 activity-independent role of HRD1 in the regulation of N-glycosylation.
Figures




Similar articles
-
Coexpression of ATP-binding cassette proteins ABCG5 and ABCG8 permits their transport to the apical surface.J Clin Invest. 2002 Sep;110(5):659-69. doi: 10.1172/JCI16000. J Clin Invest. 2002. PMID: 12208867 Free PMC article.
-
Evolutionary origin and sequence signatures of the heterodimeric ABCG5/ABCG8 transporter.Protein Sci. 2022 May;31(5):e4297. doi: 10.1002/pro.4297. Protein Sci. 2022. PMID: 35481657 Free PMC article.
-
Molecular mechanisms of subcellular localization of ABCG5 and ABCG8.Biosci Biotechnol Biochem. 2009 Mar 23;73(3):619-26. doi: 10.1271/bbb.80694. Epub 2009 Mar 7. Biosci Biotechnol Biochem. 2009. PMID: 19270375
-
ABCG5/ABCG8 in cholesterol excretion and atherosclerosis.Clin Chim Acta. 2014 Jan 20;428:82-8. doi: 10.1016/j.cca.2013.11.010. Epub 2013 Nov 16. Clin Chim Acta. 2014. PMID: 24252657 Review.
-
Role of ABCG1 and other ABCG family members in lipid metabolism.J Lipid Res. 2001 Oct;42(10):1513-20. J Lipid Res. 2001. PMID: 11590207 Review.
Cited by
-
Glaucomatous aqueous humor vesicles are smaller and differ in composition compared to controls.Exp Eye Res. 2023 Sep;234:109562. doi: 10.1016/j.exer.2023.109562. Epub 2023 Jun 27. Exp Eye Res. 2023. PMID: 37385533 Free PMC article.
-
Sitosterolemia: Twenty Years of Discovery of the Function of ABCG5ABCG8.Int J Mol Sci. 2021 Mar 5;22(5):2641. doi: 10.3390/ijms22052641. Int J Mol Sci. 2021. PMID: 33807969 Free PMC article. Review.
-
Ubiquitination in lipid metabolism reprogramming: implications for pediatric solid tumors.Front Immunol. 2025 Apr 30;16:1554311. doi: 10.3389/fimmu.2025.1554311. eCollection 2025. Front Immunol. 2025. PMID: 40370434 Free PMC article. Review.
-
Regulation of the Axillary Osmidrosis-Associated ABCC11 Protein Stability by N-Linked Glycosylation: Effect of Glucose Condition.PLoS One. 2016 Jun 9;11(6):e0157172. doi: 10.1371/journal.pone.0157172. eCollection 2016. PLoS One. 2016. PMID: 27281343 Free PMC article.
-
Identification of Glycosylation Sites Essential for Surface Expression of the CaVα2δ1 Subunit and Modulation of the Cardiac CaV1.2 Channel Activity.J Biol Chem. 2016 Feb 26;291(9):4826-43. doi: 10.1074/jbc.M115.692178. Epub 2016 Jan 7. J Biol Chem. 2016. PMID: 26742847 Free PMC article.
References
-
- Braakman I. & Bulleid N. J. Protein folding and modification in the mammalian endoplasmic reticulum. Annu. Rev. Biochem. 80, 71–99 (2011). - PubMed
-
- Helenius A. & Aebi M. Roles of N-linked glycans in the endoplasmic reticulum. Annu. Rev. Biochem. 73, 1019–1049 (2004). - PubMed
-
- Welply J. K., Shenbagamurthi P., Lennarz W. J. & Naider F. Substrate recognition by oligosaccharyltransferase. Studies on glycosylation of modified Asn-X-Thr/Ser tripeptides. J. Biol. Chem. 258, 11856–11863 (1983). - PubMed
-
- Whitley P., Nilsson I. M. & von Heijne G. A nascent secretory protein may traverse the ribosome/endoplasmic reticulum translocase complex as an extended chain. J. Biol. Chem. 271, 6241–6244 (1996). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

