CEBPA-dependent HK3 and KLF5 expression in primary AML and during AML differentiation
- PMID: 24584857
- PMCID: PMC3939455
- DOI: 10.1038/srep04261
CEBPA-dependent HK3 and KLF5 expression in primary AML and during AML differentiation
Abstract
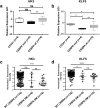
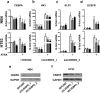
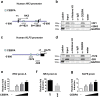
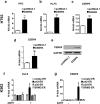
The basic leucine zipper transcription factor CCAAT/enhancer binding protein alpha (CEBPA) codes for a critical regulator during neutrophil differentiation. Aberrant expression or function of this protein contributes to the development of acute myeloid leukemia (AML). In this study, we identified two novel unrelated CEBPA target genes, the glycolytic enzyme hexokinase 3 (HK3) and the krüppel-like factor 5 (KLF5) transcription factor, by comparing gene profiles in two cohorts of CEBPA wild-type and mutant AML patients. In addition, we found CEBPA-dependent activation of HK3 and KLF5 transcription during all-trans retinoic acid (ATRA) mediated neutrophil differentiation of acute promyelocytic leukemia (APL) cells. Moreover, we observed direct regulation of HK3 by CEBPA, whereas our data suggest an indirect regulation of KLF5 by this transcription factor. Altogether, our data provide an explanation for low HK3 and KLF5 expression in particular AML subtype and establish these genes as novel CEBPA targets during neutrophil differentiation.
Figures
Similar articles
-
PU.1 is linking the glycolytic enzyme HK3 in neutrophil differentiation and survival of APL cells.Blood. 2012 May 24;119(21):4963-70. doi: 10.1182/blood-2011-09-378117. Epub 2012 Apr 12. Blood. 2012. PMID: 22498738 Free PMC article.
-
Expression and regulation of C/EBPα in normal myelopoiesis and in malignant transformation.Blood. 2017 Apr 13;129(15):2083-2091. doi: 10.1182/blood-2016-09-687822. Epub 2017 Feb 8. Blood. 2017. PMID: 28179278 Review.
-
Inactivation of the p53-KLF4-CEBPA Axis in Acute Myeloid Leukemia.Clin Cancer Res. 2016 Feb 1;22(3):746-56. doi: 10.1158/1078-0432.CCR-15-1054. Epub 2015 Sep 25. Clin Cancer Res. 2016. PMID: 26408402
-
Hexokinase 3 enhances myeloid cell survival via non-glycolytic functions.Cell Death Dis. 2022 May 11;13(5):448. doi: 10.1038/s41419-022-04891-w. Cell Death Dis. 2022. PMID: 35538058 Free PMC article.
-
Complexity of CEBPA dysregulation in human acute myeloid leukemia.Clin Cancer Res. 2009 Sep 1;15(17):5303-7. doi: 10.1158/1078-0432.CCR-08-2941. Epub 2009 Aug 25. Clin Cancer Res. 2009. PMID: 19706798 Review.
Cited by
-
HK3 is correlated with immune infiltrates and predicts response to immunotherapy in non-small cell lung cancer.Clin Transl Med. 2020 Jan;10(1):319-330. doi: 10.1002/ctm2.6. Clin Transl Med. 2020. PMID: 32508023 Free PMC article.
-
Transcriptomic Signatures and Functional Network Analysis of Chronic Rhinosinusitis With Nasal Polyps.Front Genet. 2021 Feb 2;12:609754. doi: 10.3389/fgene.2021.609754. eCollection 2021. Front Genet. 2021. PMID: 33603773 Free PMC article.
-
Regulation of emergency granulopoiesis during infection.Front Immunol. 2022 Sep 5;13:961601. doi: 10.3389/fimmu.2022.961601. eCollection 2022. Front Immunol. 2022. PMID: 36148240 Free PMC article. Review.
-
Mouse strain-specific polymorphic provirus functions as cis-regulatory element leading to epigenomic and transcriptomic variations.Nat Commun. 2021 Nov 9;12(1):6462. doi: 10.1038/s41467-021-26630-z. Nat Commun. 2021. PMID: 34753915 Free PMC article.
-
HK3 overexpression associated with epithelial-mesenchymal transition in colorectal cancer.BMC Genomics. 2018 Feb 9;19(Suppl 3):113. doi: 10.1186/s12864-018-4477-4. BMC Genomics. 2018. PMID: 29504907 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

