Evolutionary origin of higher-order repeat structure in alpha-satellite DNA of primate centromeres
- PMID: 24585002
- PMCID: PMC4131833
- DOI: 10.1093/dnares/dsu005
Evolutionary origin of higher-order repeat structure in alpha-satellite DNA of primate centromeres
Abstract
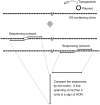
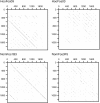
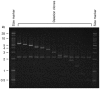
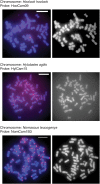
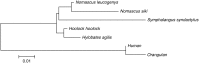
Alpha-satellite DNA (AS) is a main DNA component of primate centromeres, consisting of tandemly repeated units of ~170 bp. The AS of humans contains sequences organized into higher-order repeat (HOR) structures, in which a block of multiple repeat units forms a larger repeat unit and the larger units are repeated tandemly. The presence of HOR in AS is widely thought to be unique to hominids (family Hominidae; humans and great apes). Recently, we have identified an HOR-containing AS in the siamang, which is a small ape species belonging to the genus Symphalangus in the family Hylobatidae. This result supports the view that HOR in AS is an attribute of hominoids (superfamily Hominoidea) rather than hominids. A single example is, however, not sufficient for discussion of the evolutionary origin of HOR-containing AS. In the present study, we developed an efficient method for detecting signs of large-scale HOR and demonstrated HOR of AS in all the three other genera. Thus, AS organized into HOR occurs widely in hominoids. Our results indicate that (i) HOR-containing AS was present in the last common ancestor of hominoids or (ii) HOR-containing AS emerged independently in most or all basal branches of hominoids. We have also confirmed HOR occurrence in centromeric AS in the Hylobatidae family, which remained unclear in our previous study because of the existence of AS in subtelomeric regions, in addition to centromeres, of siamang chromosomes.
Keywords: alpha-satellite DNA; centromere; evolutionary origin; higher-order repeat; hominoids.
© The Author 2014. Published by Oxford University Press on behalf of Kazusa DNA Research Institute.
Figures

Similar articles
-
Higher-order repeat structure in alpha satellite DNA is an attribute of hominoids rather than hominids.J Hum Genet. 2013 Nov;58(11):752-4. doi: 10.1038/jhg.2013.87. Epub 2013 Aug 15. J Hum Genet. 2013. PMID: 23945983
-
Higher-order repeat structure in alpha satellite DNA occurs in New World monkeys and is not confined to hominoids.Sci Rep. 2015 May 14;5:10315. doi: 10.1038/srep10315. Sci Rep. 2015. PMID: 25974220 Free PMC article.
-
Repetitive sequences originating from the centromere constitute large-scale heterochromatin in the telomere region in the siamang, a small ape.Heredity (Edinb). 2012 Sep;109(3):180-7. doi: 10.1038/hdy.2012.28. Epub 2012 Jun 6. Heredity (Edinb). 2012. PMID: 22669075 Free PMC article.
-
Key-string algorithm--novel approach to computational analysis of repetitive sequences in human centromeric DNA.Croat Med J. 2003 Aug;44(4):386-406. Croat Med J. 2003. PMID: 12950141 Review.
-
Satellite DNAs between selfishness and functionality: structure, genomics and evolution of tandem repeats in centromeric (hetero)chromatin.Gene. 2008 Feb 15;409(1-2):72-82. doi: 10.1016/j.gene.2007.11.013. Epub 2007 Dec 4. Gene. 2008. PMID: 18182173 Review.
Cited by
-
Reduction in the structural instability of cloned eukaryotic tandem-repeat DNA by low-temperature culturing of host bacteria.Genet Res (Camb). 2014 Oct 27;96:e13. doi: 10.1017/S0016672314000172. Genet Res (Camb). 2014. PMID: 25578068 Free PMC article.
-
Locational diversity of alpha satellite DNA and intergeneric hybridization aspects in the Nomascus and Hylobates genera of small apes.PLoS One. 2014 Oct 7;9(10):e109151. doi: 10.1371/journal.pone.0109151. eCollection 2014. PLoS One. 2014. PMID: 25290445 Free PMC article.
-
High-throughput analysis of the satellitome revealed enormous diversity of satellite DNAs in the neo-Y chromosome of the cricket Eneoptera surinamensis.Sci Rep. 2017 Jul 25;7(1):6422. doi: 10.1038/s41598-017-06822-8. Sci Rep. 2017. PMID: 28743997 Free PMC article.
-
Satellite DNA: An Evolving Topic.Genes (Basel). 2017 Sep 18;8(9):230. doi: 10.3390/genes8090230. Genes (Basel). 2017. PMID: 28926993 Free PMC article. Review.
-
Novel Cascade Alpha Satellite HORs in Orangutan Chromosome 13 Assembly: Discovery of the 59mer HOR-The largest Unit in Primates-And the Missing Triplet 45/27/18 HOR in Human T2T-CHM13v2.0 Assembly.Int J Mol Sci. 2024 Jul 11;25(14):7596. doi: 10.3390/ijms25147596. Int J Mol Sci. 2024. PMID: 39062839 Free PMC article.
References
-
- Willard H.F. Evolution of alpha satellite. Curr. Opin. Genet. Dev. 1991;1:509–14. - PubMed
-
- Alexandrov I., Kazakov A., Tumeneva I., Shepelev V., Yurov Y. Alpha-satellite DNA of primates: old and new families. Chromosoma. 2001;110:253–66. - PubMed
-
- Rudd M.K., Willard H.F. Analysis of the centromeric regions of the human genome assembly. Trends Genet. 2004;20:529–33. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

