STENOFOLIA recruits TOPLESS to repress ASYMMETRIC LEAVES2 at the leaf margin and promote leaf blade outgrowth in Medicago truncatula
- PMID: 24585835
- PMCID: PMC3967031
- DOI: 10.1105/tpc.113.121947
STENOFOLIA recruits TOPLESS to repress ASYMMETRIC LEAVES2 at the leaf margin and promote leaf blade outgrowth in Medicago truncatula
Abstract
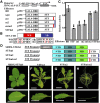
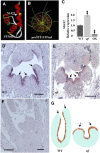
The Medicago truncatula WUSCHEL-related homeobox (WOX) gene, STENOFOLIA (STF), plays a key role in leaf blade outgrowth by promoting cell proliferation at the adaxial-abaxial junction. STF functions primarily as a transcriptional repressor, but the underlying molecular mechanism is unknown. Here, we report the identification of a protein interaction partner and a direct target, shedding light on the mechanism of STF function. Two highly conserved motifs in the C-terminal domain of STF, the WUSCHEL (WUS) box and the STF box, cooperatively recruit TOPLESS (Mt-TPL) family corepressors, and this recruitment is required for STF function, as deletion of these two domains (STFdel) impaired blade outgrowth whereas fusing Mt-TPL to STFdel restored function. The homeodomain motif is required for direct repression of ASYMMETRIC LEAVES2 (Mt-AS2), silencing of which partially rescues the stf mutant phenotype. STF and LAMINALESS1 (LAM1) are functional orthologs. A single amino acid (Asn to Ile) substitution in the homeodomain abolished the repression of Mt-AS2 and STF's ability to complement the lam1 mutant of Nicotiana sylvestris. Our data together support a model in which STF recruits corepressors to transcriptionally repress its targets during leaf blade morphogenesis. We propose that recruitment of TPL/TPL-related proteins may be a common mechanism in the repressive function of modern/WUS clade WOX genes.
Figures






References
-
- Asai T., Tena G., Plotnikova J., Willmann M.R., Chiu W.L., Gomez-Gomez L., Boller T., Ausubel F.M., Sheen J. (2002). MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415: 977–983. - PubMed
-
- Bowman J.L., Eshed Y., Baum S.F. (2002). Establishment of polarity in angiosperm lateral organs. Trends Genet. 18: 134–141. - PubMed
-
- Breuninger H., Rikirsch E., Hermann M., Ueda M., Laux T. (2008). Differential expression of WOX genes mediates apical-basal axis formation in the Arabidopsis embryo. Dev. Cell 14: 867–876. - PubMed
-
- Byrne M.E. (2012). Making leaves. Curr. Opin. Plant Biol. 15: 24–30. - PubMed
-
- Byrne M.E., Barley R., Curtis M., Arroyo J.M., Dunham M., Hudson A., Martienssen R.A. (2000). Asymmetric leaves1 mediates leaf patterning and stem cell function in Arabidopsis. Nature 408: 967–971. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

