Differential codon adaptation between dsDNA and ssDNA phages in Escherichia coli
- PMID: 24586046
- PMCID: PMC4032129
- DOI: 10.1093/molbev/msu087
Differential codon adaptation between dsDNA and ssDNA phages in Escherichia coli
Abstract
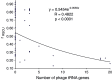
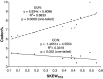
Because phages use their host translation machinery, their codon usage should evolve toward that of highly expressed host genes. We used two indices to measure codon adaptation of phages to their host, rRSCU (the correlation in relative synonymous codon usage [RSCU] between phages and their host) and Codon Adaptation Index (CAI) computed with highly expressed host genes as the reference set (because phage translation depends on host translation machinery). These indices used for this purpose are appropriate only when hosts exhibit little mutation bias, so only phages parasitizing Escherichia coli were included in the analysis. For double-stranded DNA (dsDNA) phages, both r(RSCU) and CAI decrease with increasing number of transfer RNA genes encoded by the phage genome. r(RSCU) is greater for dsDNA phages than for single-stranded DNA (ssDNA) phages, and the low r(RSCU) values are mainly due to poor concordance in RSCU values for Y-ending codons between ssDNA phages and the E. coli host, consistent with the predicted effect of C→T mutation bias in the ssDNA phages. Strong C→T mutation bias would improve codon adaptation in codon families (e.g., Gly) where U-ending codons are favored over C-ending codons ("U-friendly" codon families) by highly expressed host genes but decrease codon adaptation in other codon families where highly expressed host genes favor C-ending codons against U-ending codons ("U-hostile" codon families). It is remarkable that ssDNA phages with increasing C→T mutation bias also increased the usage of codons in the "U-friendly" codon families, thereby achieving CAI values almost as large as those of dsDNA phages. This represents a new type of codon adaptation.
Keywords: Escherichia coli; bacteriophage; codon adaptation; deamination; mutation bias; phage-host coevolution.
© The Author 2014. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures

References
-
- Azeredo J, Sutherland IW. The use of phages for the removal of infectious biofilms. Curr Pharm Biotechnol. 2008;9:261–266. - PubMed
-
- Brussow H, Kutter E. Genomics and evolution of tailed phages. In: Kutter E, Sulakvelidze A, editors. Bacteriophages: biology and applications. Boca Raton (FL): CRC Press; 2005. pp. 91–128.
-
- Bulmer M. Coevolution of codon usage and transfer RNA abundance. Nature. 1987;325:728–730. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

