A combination of transcriptional and microRNA regulation improves the stability of the relative concentrations of target genes
- PMID: 24586138
- PMCID: PMC3937125
- DOI: 10.1371/journal.pcbi.1003490
A combination of transcriptional and microRNA regulation improves the stability of the relative concentrations of target genes
Erratum in
- PLoS Comput Biol. 2014 Mar;10(3):e1003582
Abstract
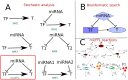
It is well known that, under suitable conditions, microRNAs are able to fine tune the relative concentration of their targets to any desired value. We show that this function is particularly effective when one of the targets is a Transcription Factor (TF) which regulates the other targets. This combination defines a new class of feed-forward loops (FFLs) in which the microRNA plays the role of master regulator. Using both deterministic and stochastic equations, we show that these FFLs are indeed able not only to fine-tune the TF/target ratio to any desired value as a function of the miRNA concentration but also, thanks to the peculiar topology of the circuit, to ensure the stability of this ratio against stochastic fluctuations. These two effects are due to the interplay between the direct transcriptional regulation and the indirect TF/Target interaction due to competition of TF and target for miRNA binding (the so called "sponge effect"). We then perform a genome wide search of these FFLs in the human regulatory network and show that they are characterized by a very peculiar enrichment pattern. In particular, they are strongly enriched in all the situations in which the TF and its target have to be precisely kept at the same concentration notwithstanding the environmental noise. As an example we discuss the FFL involving E2F1 as Transcription Factor, RB1 as target and miR-17 family as master regulator. These FFLs ensure a tight control of the E2F/RB ratio which in turns ensures the stability of the transition from the G0/G1 to the S phase in quiescent cells.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
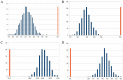

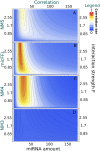
 is mapped as a function of the miRNA concentration and of the interaction strength
is mapped as a function of the miRNA concentration and of the interaction strength  . While for NM3 and NM5 the fluctuation of TF and T are almost uncorrelated, both NM4 and the micFFL show a well defined region of large correlation. This correlation occurs for rather low miRNA concentrations and for almost any value of the miRNA-mRNA interaction strength.
. While for NM3 and NM5 the fluctuation of TF and T are almost uncorrelated, both NM4 and the micFFL show a well defined region of large correlation. This correlation occurs for rather low miRNA concentrations and for almost any value of the miRNA-mRNA interaction strength.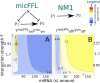
References
-
- Alon U (2007) Network motifs: theory and experimental approaches. Nature Rev Genet 8: 450–461. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

