The post-transcriptional regulator rsmA/csrA activates T3SS by stabilizing the 5' UTR of hrpG, the master regulator of hrp/hrc genes, in Xanthomonas
- PMID: 24586158
- PMCID: PMC3937308
- DOI: 10.1371/journal.ppat.1003945
The post-transcriptional regulator rsmA/csrA activates T3SS by stabilizing the 5' UTR of hrpG, the master regulator of hrp/hrc genes, in Xanthomonas
Abstract
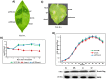
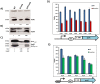
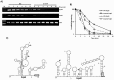
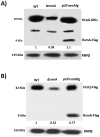
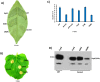
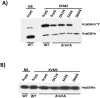
The RsmA/CsrA family of the post-transcriptional regulators of bacteria is involved in the regulation of many cellular processes, including pathogenesis. In this study, we demonstrated that rsmA not only is required for the full virulence of the phytopathogenic bacterium Xanthomonas citri subsp. citri (XCC) but also contributes to triggering the hypersensitive response (HR) in non-host plants. Deletion of rsmA resulted in significantly reduced virulence in the host plant sweet orange and a delayed and weakened HR in the non-host plant Nicotiana benthamiana. Microarray, quantitative reverse-transcription PCR, western-blotting, and GUS assays indicated that RsmA regulates the expression of the type 3 secretion system (T3SS) at both transcriptional and post-transcriptional levels. The regulation of T3SS by RsmA is a universal phenomenon in T3SS-containing bacteria, but the specific mechanism seems to depend on the interaction between a particular bacterium and its hosts. For Xanthomonads, the mechanism by which RsmA activates T3SS remains unknown. Here, we show that RsmA activates the expression of T3SS-encoding hrp/hrc genes by directly binding to the 5' untranslated region (UTR) of hrpG, the master regulator of the hrp/hrc genes in XCC. RsmA stabilizes hrpG mRNA, leading to increased accumulation of HrpG proteins and subsequently, the activation of hrp/hrc genes. The activation of the hrp/hrc genes by RsmA via HrpG was further supported by the observation that ectopic overexpression of hrpG in an rsmA mutant restored its ability to cause disease in host plants and trigger HR in non-host plants. RsmA also stabilizes the transcripts of another T3SS-associated hrpD operon by directly binding to the 5' UTR region. Taken together, these data revealed that RsmA primarily activates T3SS by acting as a positive regulator of hrpG and that this regulation is critical to the pathogenicity of XCC.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
-
- Buttner D, Bonas U (2010) Regulation and secretion of Xanthomonas virulence factors. FEMS microbiology reviews 34: 107–133. - PubMed
-
- Kay S, Bonas U (2009) How Xanthomonas type III effectors manipulate the host plant. Current opinion in microbiology 12: 37–43. - PubMed
-
- Boch J, Bonas U (2010) Xanthomonas AvrBs3 family-type III effectors: discovery and function. Annual review of phytopathology 48: 419–436. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

