A variant in the neuropeptide receptor npr-1 is a major determinant of Caenorhabditis elegans growth and physiology
- PMID: 24586193
- PMCID: PMC3937155
- DOI: 10.1371/journal.pgen.1004156
A variant in the neuropeptide receptor npr-1 is a major determinant of Caenorhabditis elegans growth and physiology
Erratum in
- PLoS Genet. 2014 Mar;10(3):e1004316
Abstract
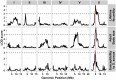
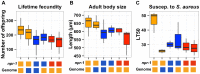
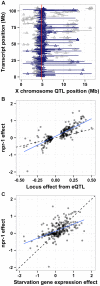
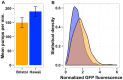
The mechanistic basis for how genetic variants cause differences in phenotypic traits is often elusive. We identified a quantitative trait locus in Caenorhabditis elegans that affects three seemingly unrelated phenotypic traits: lifetime fecundity, adult body size, and susceptibility to the human pathogen Staphyloccus aureus. We found a QTL for all three traits arises from variation in the neuropeptide receptor gene npr-1. Moreover, we found that variation in npr-1 is also responsible for differences in 247 gene expression traits. Variation in npr-1 is known to determine whether animals disperse throughout a bacterial lawn or aggregate at the edges of the lawn. We found that the allele that leads to aggregation is associated with reduced growth and reproductive output. The altered gene expression pattern caused by this allele suggests that the aggregation behavior might cause a weak starvation state, which is known to reduce growth rate and fecundity. Importantly, we show that variation in npr-1 causes each of these phenotypic differences through behavioral avoidance of ambient oxygen concentrations. These results suggest that variation in npr-1 has broad pleiotropic effects mediated by altered exposure to bacterial food.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
-
- Rockman MV, Skrovanek SS, Kruglyak L (2010) Selection at linked sites shapes heritable phenotypic variation in C. elegans. Science 330: 372–376 doi:10.1126/science.1194208 - DOI - PMC - PubMed
-
- Seidel HS, Ailion M, Li J, van Oudenaarden A, Rockman MV, et al. (2011) A novel sperm-delivered toxin causes late-stage embryo lethality and transmission ratio distortion in C. elegans. Plos Biol 9: e1001115 doi:10.1371/journal.pbio.1001115 - DOI - PMC - PubMed
-
- Seidel HS, Rockman MV, Kruglyak L (2008) Widespread genetic incompatibility in C. elegans maintained by balancing selection. Science 319: 589–594 doi:10.1126/science.1151107 - DOI - PMC - PubMed
-
- Ghosh R, Andersen EC, Shapiro JA, Gerke JP, Kruglyak L (2012) Natural variation in a chloride channel subunit confers avermectin resistance in C. elegans. Science 335: 574–578 doi:10.1126/science.1214318 - DOI - PMC - PubMed
-
- Bendesky A, Tsunozaki M, Rockman MV, Kruglyak L, Bargmann CI (2011) Catecholamine receptor polymorphisms affect decision-making in C. elegans. Nature 472: 313–318 doi:10.1038/nature09821 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- F32-GM089007/GM/NIGMS NIH HHS/United States
- P30 CA060553/CA/NCI NIH HHS/United States
- T32 GM007388/GM/NIGMS NIH HHS/United States
- T32 CA009528/CA/NCI NIH HHS/United States
- T32-CA009528/CA/NCI NIH HHS/United States
- F32 GM089007/GM/NIGMS NIH HHS/United States
- R37-MH59520/MH/NIMH NIH HHS/United States
- R37 MH059520/MH/NIMH NIH HHS/United States
- P50-GM071508/GM/NIGMS NIH HHS/United States
- R01 HG004321/HG/NHGRI NIH HHS/United States
- P50 GM071508/GM/NIGMS NIH HHS/United States
- R01-HG004321/HG/NHGRI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

