DAAM is required for thin filament formation and Sarcomerogenesis during muscle development in Drosophila
- PMID: 24586196
- PMCID: PMC3937221
- DOI: 10.1371/journal.pgen.1004166
DAAM is required for thin filament formation and Sarcomerogenesis during muscle development in Drosophila
Abstract
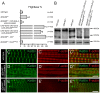
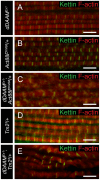
During muscle development, myosin and actin containing filaments assemble into the highly organized sarcomeric structure critical for muscle function. Although sarcomerogenesis clearly involves the de novo formation of actin filaments, this process remained poorly understood. Here we show that mouse and Drosophila members of the DAAM formin family are sarcomere-associated actin assembly factors enriched at the Z-disc and M-band. Analysis of dDAAM mutants revealed a pivotal role in myofibrillogenesis of larval somatic muscles, indirect flight muscles and the heart. We found that loss of dDAAM function results in multiple defects in sarcomere development including thin and thick filament disorganization, Z-disc and M-band formation, and a near complete absence of the myofibrillar lattice. Collectively, our data suggest that dDAAM is required for the initial assembly of thin filaments, and subsequently it promotes filament elongation by assembling short actin polymers that anneal to the pointed end of the growing filaments, and by antagonizing the capping protein Tropomodulin.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





Similar articles
-
Troponin I is required for myofibrillogenesis and sarcomere formation in Drosophila flight muscle.J Cell Sci. 2004 Apr 1;117(Pt 9):1795-805. doi: 10.1242/jcs.01024. Epub 2004 Mar 16. J Cell Sci. 2004. PMID: 15075240
-
Thin filaments elongate from their pointed ends during myofibril assembly in Drosophila indirect flight muscle.J Cell Biol. 2001 Dec 10;155(6):1043-53. doi: 10.1083/jcb.200108026. Epub 2001 Dec 10. J Cell Biol. 2001. PMID: 11739412 Free PMC article.
-
Characterization of the biochemical properties and biological function of the formin homology domains of Drosophila DAAM.J Biol Chem. 2010 Apr 23;285(17):13154-69. doi: 10.1074/jbc.M109.093914. Epub 2010 Feb 21. J Biol Chem. 2010. PMID: 20177055 Free PMC article.
-
Tropomodulin capping of actin filaments in striated muscle development and physiology.J Biomed Biotechnol. 2011;2011:103069. doi: 10.1155/2011/103069. Epub 2011 Oct 17. J Biomed Biotechnol. 2011. PMID: 22013379 Free PMC article. Review.
-
Thin filament length regulation in striated muscle sarcomeres: pointed-end dynamics go beyond a nebulin ruler.Semin Cell Dev Biol. 2008 Dec;19(6):511-9. doi: 10.1016/j.semcdb.2008.08.009. Epub 2008 Aug 26. Semin Cell Dev Biol. 2008. PMID: 18793739 Free PMC article. Review.
Cited by
-
Assembly and Maintenance of Sarcomere Thin Filaments and Associated Diseases.Int J Mol Sci. 2020 Jan 15;21(2):542. doi: 10.3390/ijms21020542. Int J Mol Sci. 2020. PMID: 31952119 Free PMC article. Review.
-
The Drosophila formin Fhos is a primary mediator of sarcomeric thin-filament array assembly.Elife. 2016 Oct 12;5:e16540. doi: 10.7554/eLife.16540. Elife. 2016. PMID: 27731794 Free PMC article.
-
The Drosophila actin nucleator DAAM is essential for left-right asymmetry.PLoS Genet. 2020 Apr 23;16(4):e1008758. doi: 10.1371/journal.pgen.1008758. eCollection 2020 Apr. PLoS Genet. 2020. PMID: 32324733 Free PMC article.
-
Human formin FHOD3-mediated actin elongation is required for sarcomere integrity in cardiomyocytes.Elife. 2025 Jul 15;13:RP104048. doi: 10.7554/eLife.104048. Elife. 2025. PMID: 40663059 Free PMC article.
-
The actin polymerization factor Diaphanous and the actin severing protein Flightless I collaborate to regulate sarcomere size.Dev Biol. 2021 Jan 1;469:12-25. doi: 10.1016/j.ydbio.2020.09.014. Epub 2020 Sep 25. Dev Biol. 2021. PMID: 32980309 Free PMC article.
References
-
- Sparrow JC, Schock F (2009) The initial steps of myofibril assembly: integrins pave the way. Nat Rev Mol Cell Biol 10: 293–298. - PubMed
-
- Pollard TD, Blanchoin L, Mullins RD (2000) Molecular mechanisms controlling actin filament dynamics in nonmuscle cells. Annu Rev Biophys Biomol Struct 29: 545–576. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

