Chromosome I controls chromosome II replication in Vibrio cholerae
- PMID: 24586205
- PMCID: PMC3937223
- DOI: 10.1371/journal.pgen.1004184
Chromosome I controls chromosome II replication in Vibrio cholerae
Abstract
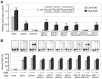
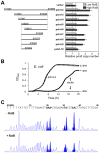
Control of chromosome replication involves a common set of regulators in eukaryotes, whereas bacteria with divided genomes use chromosome-specific regulators. How bacterial chromosomes might communicate for replication is not known. In Vibrio cholerae, which has two chromosomes (chrI and chrII), replication initiation is controlled by DnaA in chrI and by RctB in chrII. DnaA has binding sites at the chrI origin of replication as well as outside the origin. RctB likewise binds at the chrII origin and, as shown here, to external sites. The binding to the external sites in chrII inhibits chrII replication. A new kind of site was found in chrI that enhances chrII replication. Consistent with its enhancing activity, the chrI site increased RctB binding to those chrII origin sites that stimulate replication and decreased binding to other sites that inhibit replication. The differential effect on binding suggests that the new site remodels RctB. The chaperone-like activity of the site is supported by the finding that it could relieve the dependence of chrII replication on chaperone proteins DnaJ and DnaK. The presence of a site in chrI that specifically controls chrII replication suggests a mechanism for communication between the two chromosomes for replication.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
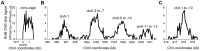


Similar articles
-
Regulatory cross-talk links Vibrio cholerae chromosome II replication and segregation.PLoS Genet. 2011 Jul;7(7):e1002189. doi: 10.1371/journal.pgen.1002189. Epub 2011 Jul 21. PLoS Genet. 2011. PMID: 21811418 Free PMC article.
-
Insensitivity of chromosome I and the cell cycle to blockage of replication and segregation of Vibrio cholerae chromosome II.mBio. 2012 May 8;3(3):e00067-12. doi: 10.1128/mBio.00067-12. Print 2012. mBio. 2012. PMID: 22570276 Free PMC article.
-
Replication regulation of Vibrio cholerae chromosome II involves initiator binding to the origin both as monomer and as dimer.Nucleic Acids Res. 2012 Jul;40(13):6026-38. doi: 10.1093/nar/gks260. Epub 2012 Mar 24. Nucleic Acids Res. 2012. PMID: 22447451 Free PMC article.
-
Cell cycle-coordinated maintenance of the Vibrio bipartite genome.EcoSal Plus. 2023 Dec 12;11(1):eesp00082022. doi: 10.1128/ecosalplus.esp-0008-2022. Epub 2023 Nov 22. EcoSal Plus. 2023. PMID: 38277776 Free PMC article. Review.
-
Random versus Cell Cycle-Regulated Replication Initiation in Bacteria: Insights from Studying Vibrio cholerae Chromosome 2.Microbiol Mol Biol Rev. 2016 Nov 30;81(1):e00033-16. doi: 10.1128/MMBR.00033-16. Print 2017 Mar. Microbiol Mol Biol Rev. 2016. PMID: 27903655 Free PMC article. Review.
Cited by
-
oriC-encoded instructions for the initiation of bacterial chromosome replication.Front Microbiol. 2015 Jan 6;5:735. doi: 10.3389/fmicb.2014.00735. eCollection 2014. Front Microbiol. 2015. PMID: 25610430 Free PMC article. Review.
-
A Requirement for Global Transcription Factor Lrp in Licensing Replication of Vibrio cholerae Chromosome 2.Front Microbiol. 2018 Sep 10;9:2103. doi: 10.3389/fmicb.2018.02103. eCollection 2018. Front Microbiol. 2018. PMID: 30250457 Free PMC article.
-
A checkpoint control orchestrates the replication of the two chromosomes of Vibrio cholerae.Sci Adv. 2016 Apr 22;2(4):e1501914. doi: 10.1126/sciadv.1501914. eCollection 2016 Apr. Sci Adv. 2016. PMID: 27152358 Free PMC article.
-
The DnaK Chaperone Uses Different Mechanisms To Promote and Inhibit Replication of Vibrio cholerae Chromosome 2.mBio. 2017 Apr 18;8(2):e00427-17. doi: 10.1128/mBio.00427-17. mBio. 2017. PMID: 28420739 Free PMC article.
-
Rationally designed chromosome fusion does not prevent rapid growth of Vibrio natriegens.Commun Biol. 2024 May 2;7(1):519. doi: 10.1038/s42003-024-06234-1. Commun Biol. 2024. PMID: 38698198 Free PMC article.
References
-
- Harrison PW, Lower RP, Kim NK, Young JP (2010) Introducing the bacterial ‘chromid’: not a chromosome, not a plasmid. Trends Microbiol 18: 141–148. - PubMed
-
- Kawakami H, Katayama T (2010) DnaA, ORC, and Cdc6: similarity beyond the domains of life and diversity. Biochem Cell Biol 88: 49–62. - PubMed
-
- Giraldo R (2003) Common domains in the initiators of DNA replication in Bacteria, Archaea and Eukarya: combined structural, functional and phylogenetic perspectives. FEMS Microbiol Rev 26: 533–554. - PubMed
-
- Egan ES, Fogel MA, Waldor MK (2005) Divided genomes: negotiating the cell cycle in prokaryotes with multiple chromosomes. Mol Microbiol 56: 1129–1138. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

