DNA aptamers against exon v10 of CD44 inhibit breast cancer cell migration
- PMID: 24586375
- PMCID: PMC3929491
- DOI: 10.1371/journal.pone.0088712
DNA aptamers against exon v10 of CD44 inhibit breast cancer cell migration
Abstract
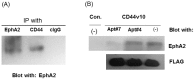
CD44 adhesion molecules are expressed in many breast cancer cells and have been demonstrated to play a key role in regulating malignant phenotypes such as growth, migration, and invasion. CD44 is an integral transmembrane protein encoded by a single 20-exon gene. The diversity of the biological functions of CD44 is the result of the various splicing variants of these exons. Previous studies suggest that exon v10 of CD44 plays a key role in promoting cancer invasion and metastasis, however, the molecular mechanisms are not clear. Given the fact that exon v10 is in the ectodomain of CD44, we hypothesized that CD44 forms a molecular complex with other cell surface molecules through exon v10 in order to promote migration of breast cancer cells. In order to test this hypothesis, we selected DNA aptamers that specifically bound to CD44 exon v10 using Systematic Evolution of Ligands by Exponential Enrichment (SELEX). We selected aptamers that inhibited migration of breast cancer cells. Co-immunoprecipitation studies demonstrated that EphA2 was co-precipitated with CD44. Pull-down studies demonstrated that recombinant CD44 exon v10 bound to EphA2 and more importantly aptamers that inhibited migration also prevented the binding of EphA2 to exon v10. These results suggest that CD44 forms a molecular complex with EphA2 on the breast cancer cell surface and this complex plays a key role in enhancing breast cancer migration. These results provide insight not only for characterizing mechanisms of breast cancer migration but also for developing target-specific therapy for breast cancers and possibly other cancer types expressing CD44 exon v10.
Conflict of interest statement
Figures




References
-
- Ferreira CS, Matthews CS, Missailidis S (2006) DNA aptamers that bind to MUC1 tumour marker: design and characterization of MUC1-binding single-stranded DNA aptamers. Tumour Biol 27: 289–301. - PubMed
-
- Fitzwater T, Polisky B (1996) A SELEX primer. Methods Enzymol 267: 275–301. - PubMed
-
- Lupold SE, Hicke BJ, Lin Y, Coffey DS (2002) Identification and characterization of nuclease-stabilized RNA molecules that bind human prostate cancer cells via the prostate-specific membrane antigen. Cancer Res 62: 4029–4033. - PubMed
-
- Kimoto M, Shirouzu M, Mizutani S, Koide H, Kaziro Y, et al. (2002) Anti-(Raf-1) RNA aptamers that inhibit Ras-induced Raf-1 activation. Eur J Biochem 269: 697–704. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

