Neuronal-specific deficiency of the splicing factor Tra2b causes apoptosis in neurogenic areas of the developing mouse brain
- PMID: 24586484
- PMCID: PMC3929626
- DOI: 10.1371/journal.pone.0089020
Neuronal-specific deficiency of the splicing factor Tra2b causes apoptosis in neurogenic areas of the developing mouse brain
Abstract
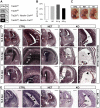
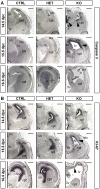
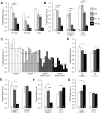
Alternative splicing (AS) increases the informational content of the genome and is more prevalent in the brain than in any other tissue. The splicing factor Tra2b (Sfrs10) can modulate splicing inclusion of exons by specifically detecting GAA-rich binding motifs and its absence causes early embryonic lethality in mice. TRA2B has been shown to be involved in splicing processes of Nasp (nuclear autoantigenic sperm protein), MAPT (microtubule associated protein tau) and SMN (survival motor neuron), and is therefore implicated in spermatogenesis and neurological diseases like Alzheimer's disease, dementia, Parkinson's disease and spinal muscular atrophy. Here we generated a neuronal-specific Tra2b knock-out mouse that lacks Tra2b expression in neuronal and glial precursor cells by using the Nestin-Cre. Neuronal-specific Tra2b knock-out mice die immediately after birth and show severe abnormalities in cortical development, which are caused by massive apoptotic events in the ventricular layers of the cortex, demonstrating a pivotal role of Tra2b for the developing central nervous system. Using whole brain RNA on exon arrays we identified differentially expressed alternative exons of Tubulinδ1 and Shugoshin-like2 as in vivo targets of Tra2b. Most interestingly, we found increased expression of the cyclin dependent kinase inhibitor 1a (p21) which we could functionally link to neuronal precursor cells in the affected brain regions. We provide further evidence that the absence of Tra2b causes p21 upregulation and ultimately cell death in NSC34 neuronal-like cells. These findings demonstrate that Tra2b regulates splicing events essential for maintaining neuronal viability during development. Apoptotic events triggered via p21 might not be restricted to the developing brain but could possibly be generalized to the whole organism and explain early embryonic lethality in Tra2b-depleted mice.
Conflict of interest statement
Figures



Similar articles
-
Splicing factor TRA2B is required for neural progenitor survival.J Comp Neurol. 2014 Feb 1;522(2):372-92. doi: 10.1002/cne.23405. J Comp Neurol. 2014. PMID: 23818142 Free PMC article.
-
Deficiency of the splicing factor Sfrs10 results in early embryonic lethality in mice and has no impact on full-length SMN/Smn splicing.Hum Mol Genet. 2010 Jun 1;19(11):2154-67. doi: 10.1093/hmg/ddq094. Epub 2010 Feb 27. Hum Mol Genet. 2010. PMID: 20190275
-
Identification of evolutionarily conserved exons as regulated targets for the splicing activator tra2β in development.PLoS Genet. 2011 Dec;7(12):e1002390. doi: 10.1371/journal.pgen.1002390. Epub 2011 Dec 15. PLoS Genet. 2011. PMID: 22194695 Free PMC article.
-
RNA splicing regulators play critical roles in neurogenesis.Wiley Interdiscip Rev RNA. 2022 Nov;13(6):e1728. doi: 10.1002/wrna.1728. Epub 2022 Apr 6. Wiley Interdiscip Rev RNA. 2022. PMID: 35388651 Review.
-
Functional and mechanistic insights from genome-wide studies of splicing regulation in the brain.Adv Exp Med Biol. 2007;623:148-60. doi: 10.1007/978-0-387-77374-2_9. Adv Exp Med Biol. 2007. PMID: 18380345 Review.
Cited by
-
The Important Role of Perituberal Tissue in Epileptic Patients with Tuberous Sclerosis Complex by the Transcriptome Analysis.Biomed Res Int. 2020 Oct 15;2020:4980609. doi: 10.1155/2020/4980609. eCollection 2020. Biomed Res Int. 2020. PMID: 33123575 Free PMC article.
-
SRC Kinase Isoforms Regulate mRNA Splicing during Neural Development.J Neurosci. 2025 Aug 20;45(34):e1705242025. doi: 10.1523/JNEUROSCI.1705-24.2025. J Neurosci. 2025. PMID: 40750357 Free PMC article.
-
Human Tra2 proteins jointly control a CHEK1 splicing switch among alternative and constitutive target exons.Nat Commun. 2014 Sep 11;5:4760. doi: 10.1038/ncomms5760. Nat Commun. 2014. PMID: 25208576 Free PMC article.
-
Alternative-splicing defects in cancer: Splicing regulators and their downstream targets, guiding the way to novel cancer therapeutics.Wiley Interdiscip Rev RNA. 2018 Jul;9(4):e1476. doi: 10.1002/wrna.1476. Epub 2018 Apr 25. Wiley Interdiscip Rev RNA. 2018. PMID: 29693319 Free PMC article. Review.
-
Poison Exon Splicing Regulates a Coordinated Network of SR Protein Expression during Differentiation and Tumorigenesis.Mol Cell. 2020 Nov 19;80(4):648-665.e9. doi: 10.1016/j.molcel.2020.10.019. Epub 2020 Nov 10. Mol Cell. 2020. PMID: 33176162 Free PMC article.
References
-
- Lareau LF, Inada M, Green RE, Wengrod JC, Brenner SE (2007) Unproductive splicing of SR genes associated with highly conserved and ultraconserved DNA elements. Nature 446: 926–929. - PubMed
-
- McGlincy NJ, Smith CW (2008) Alternative splicing resulting in nonsense-mediated mRNA decay: what is the meaning of nonsense? Trends Biochem Sci 33: 385–393. - PubMed
-
- Matlin AJ, Clark F, Smith CW (2005) Understanding alternative splicing: towards a cellular code. NatRevMolCell Biol 6: 386–398. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

