Distinct structural features of the peroxide response regulator from group A Streptococcus drive DNA binding
- PMID: 24586487
- PMCID: PMC3931707
- DOI: 10.1371/journal.pone.0089027
Distinct structural features of the peroxide response regulator from group A Streptococcus drive DNA binding
Abstract
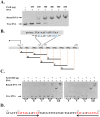
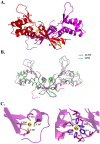
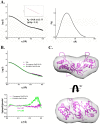
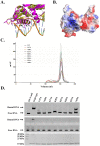
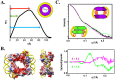
Group A streptococcus (GAS, Streptococcus pyogenes) is a strict human pathogen that causes severe, invasive diseases. GAS does not produce catalase, but has an ability to resist killing by reactive oxygen species (ROS) through novel mechanisms. The peroxide response regulator (PerR), a member of ferric uptake regulator (Fur) family, plays a key role for GAS to cope with oxidative stress by regulating the expression of multiple genes. Our previous studies have found that expression of an iron-binding protein, Dpr, is under the direct control of PerR. To elucidate the molecular interactions of PerR with its cognate promoter, we have carried out structural studies on PerR and PerR-DNA complex. By combining crystallography and small-angle X-ray scattering (SAXS), we confirmed that the determined PerR crystal structure reflects its conformation in solution. Through mutagenesis and biochemical analysis, we have identified DNA-binding residues suggesting that PerR binds to the dpr promoter at the per box through a winged-helix motif. Furthermore, we have performed SAXS analysis and resolved the molecular architecture of PerR-DNA complex, in which two 30 bp DNA fragments wrap around two PerR homodimers by interacting with the adjacent positively-charged winged-helix motifs. Overall, we provide structural insights into molecular recognition of DNA by PerR and define the hollow structural arrangement of PerR-30bpDNA complex, which displays a unique topology distinct from currently proposed DNA-binding models for Fur family regulators.
Conflict of interest statement
Figures

Similar articles
-
Crystal structure of peroxide stress regulator from Streptococcus pyogenes provides functional insights into the mechanism of oxidative stress sensing.J Biol Chem. 2013 Jun 21;288(25):18311-24. doi: 10.1074/jbc.M113.456590. Epub 2013 May 3. J Biol Chem. 2013. PMID: 23645680 Free PMC article.
-
A Fur-like protein PerR regulates two oxidative stress response related operons dpr and metQIN in Streptococcus suis.BMC Microbiol. 2012 May 30;12:85. doi: 10.1186/1471-2180-12-85. BMC Microbiol. 2012. PMID: 22646062 Free PMC article.
-
Oxidative stress and metal ions regulate a ferritin-like gene, dpr, in Streptococcus pyogenes.Int J Med Microbiol. 2010 Apr;300(4):259-64. doi: 10.1016/j.ijmm.2009.09.002. Epub 2009 Oct 29. Int J Med Microbiol. 2010. PMID: 19879189
-
Transcriptional regulation by Ferric Uptake Regulator (Fur) in pathogenic bacteria.Front Cell Infect Microbiol. 2013 Oct 2;3:59. doi: 10.3389/fcimb.2013.00059. eCollection 2013. Front Cell Infect Microbiol. 2013. PMID: 24106689 Free PMC article. Review.
-
Functional specialization within the Fur family of metalloregulators.Biometals. 2007 Jun;20(3-4):485-99. doi: 10.1007/s10534-006-9070-7. Epub 2007 Jan 10. Biometals. 2007. PMID: 17216355 Review.
Cited by
-
Structural and mechanistic basis of zinc regulation across the E. coli Zur regulon.PLoS Biol. 2014 Nov 4;12(11):e1001987. doi: 10.1371/journal.pbio.1001987. eCollection 2014 Nov. PLoS Biol. 2014. PMID: 25369000 Free PMC article.
-
BosR Is A Novel Fur Family Member Responsive to Copper and Regulating Copper Homeostasis in Borrelia burgdorferi.J Bacteriol. 2017 Jul 25;199(16):e00276-17. doi: 10.1128/JB.00276-17. Print 2017 Aug 15. J Bacteriol. 2017. PMID: 28583949 Free PMC article.
-
Structural basis for zinc-induced activation of a zinc uptake transcriptional regulator.Nucleic Acids Res. 2021 Jun 21;49(11):6511-6528. doi: 10.1093/nar/gkab432. Nucleic Acids Res. 2021. PMID: 34048589 Free PMC article.
-
Structural basis for persulfide-sensing specificity in a transcriptional regulator.Nat Chem Biol. 2021 Jan;17(1):65-70. doi: 10.1038/s41589-020-00671-9. Epub 2020 Oct 26. Nat Chem Biol. 2021. PMID: 33106663 Free PMC article.
-
The CXXC Motifs Are Essential for the Function of BosR in Borrelia burgdorferi.Front Cell Infect Microbiol. 2019 Apr 16;9:109. doi: 10.3389/fcimb.2019.00109. eCollection 2019. Front Cell Infect Microbiol. 2019. PMID: 31041197 Free PMC article.
References
-
- Carapetis JR, Steer AC, Mulholland EK, Weber M (2005) The global burden of group A streptococcal diseases. Lancet Infect Dis 5: 685–694. - PubMed
-
- Troillet N LA, de Werra P, Praz G (1994) Invasive Streptococcus pyogenes infection (P-hemolytic streptococcus of group A). Schweiz Med Wochenschr 124: 1064–1069. - PubMed
-
- Brenot A, King KY, Caparon MG (2005) The PerR regulon in peroxide resistance and virulence of Streptococcus pyogenes . Mol Microbiol 55: 221–234. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

