Discovery of selective inhibitors of the Clostridium difficile dehydroquinate dehydratase
- PMID: 24586713
- PMCID: PMC3931744
- DOI: 10.1371/journal.pone.0089356
Discovery of selective inhibitors of the Clostridium difficile dehydroquinate dehydratase
Abstract
A vibrant and healthy gut flora is essential for preventing the proliferation of Clostridium difficile, a pathogenic bacterium that causes severe gastrointestinal symptoms. In fact, most C. difficile infections (CDIs) occur after broad-spectrum antibiotic treatment, which, by eradicating the commensal gut bacteria, allows its spores to proliferate. Hence, a C. difficile specific antibiotic that spares the gut flora would be highly beneficial in treating CDI. Towards this goal, we set out to discover small molecule inhibitors of the C. difficile enzyme dehydroquinate dehydratase (DHQD). DHQD is the 3(rd) of seven enzymes that compose the shikimate pathway, a metabolic pathway absent in humans, and is present in bacteria as two phylogenetically and mechanistically distinct types. Using a high-throughput screen we identified three compounds that inhibited the type I C. difficile DHQD but not the type II DHQD from Bacteroides thetaiotaomicron, a highly represented commensal gut bacterial species. Kinetic analysis revealed that the compounds inhibit the C. difficile enzyme with Ki values ranging from 10 to 20 µM. Unexpectedly, kinetic and biophysical studies demonstrate that inhibitors also exhibit selectivity between type I DHQDs, inhibiting the C. difficile but not the highly homologous Salmonella enterica DHQD. Therefore, the three identified compounds seem to be promising lead compounds for the development of C. difficile specific antibiotics.
Conflict of interest statement
Figures
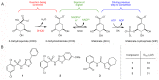

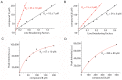
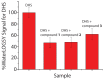
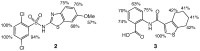
Similar articles
-
Clostridium difficile drug pipeline: challenges in discovery and development of new agents.J Med Chem. 2015 Jul 9;58(13):5164-85. doi: 10.1021/jm5016846. Epub 2015 Mar 30. J Med Chem. 2015. PMID: 25760275 Free PMC article. Review.
-
Insights into the mechanism of type I dehydroquinate dehydratases from structures of reaction intermediates.J Biol Chem. 2011 Feb 4;286(5):3531-9. doi: 10.1074/jbc.M110.192831. Epub 2010 Nov 18. J Biol Chem. 2011. PMID: 21087925 Free PMC article.
-
A high-throughput small-molecule screen to identify a novel chemical inhibitor of Clostridium difficile.Int J Antimicrob Agents. 2014 Jul;44(1):69-73. doi: 10.1016/j.ijantimicag.2014.03.007. Epub 2014 Apr 24. Int J Antimicrob Agents. 2014. PMID: 24837414 Free PMC article.
-
Development of machine learning models to predict inhibition of 3-dehydroquinate dehydratase.Chem Biol Drug Des. 2018 Aug;92(2):1468-1474. doi: 10.1111/cbdd.13312. Epub 2018 May 18. Chem Biol Drug Des. 2018. PMID: 29676519
-
Advances in the treatment of Clostridium difficile with fidaxomicin: a narrow spectrum antibiotic.Ann N Y Acad Sci. 2013 Jul;1291:33-41. doi: 10.1111/nyas.12135. Epub 2013 May 14. Ann N Y Acad Sci. 2013. PMID: 23672600 Review.
Cited by
-
Clostridium difficile drug pipeline: challenges in discovery and development of new agents.J Med Chem. 2015 Jul 9;58(13):5164-85. doi: 10.1021/jm5016846. Epub 2015 Mar 30. J Med Chem. 2015. PMID: 25760275 Free PMC article. Review.
-
Transmissible microbial and metabolomic remodeling by soluble dietary fiber improves metabolic homeostasis.Sci Rep. 2015 Jun 4;5:10604. doi: 10.1038/srep10604. Sci Rep. 2015. PMID: 26040234 Free PMC article.
-
Discovery of Potential Noncovalent Inhibitors of Dehydroquinate Dehydratase from Methicillin-Resistant Staphylococcus aureus through Computational-Driven Drug Design.Pharmaceuticals (Basel). 2023 Aug 12;16(8):1148. doi: 10.3390/ph16081148. Pharmaceuticals (Basel). 2023. PMID: 37631063 Free PMC article.
References
-
- Rupnik M, Wilcox MH, Gerding DN (2009) Clostridium difficile infection: new developments in epidemiology and pathogenesis. Nat Rev Microbiol 7: 526–536. - PubMed
-
- Kuijper EJ, van Dissel JT, Wilcox MH (2007) Clostridium difficile: changing epidemiology and new treatment options. Curr Opin Infect Dis 20: 376–383. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

