Effect of bacteria on the wound healing behavior of oral epithelial cells
- PMID: 24586806
- PMCID: PMC3931835
- DOI: 10.1371/journal.pone.0089475
Effect of bacteria on the wound healing behavior of oral epithelial cells
Abstract
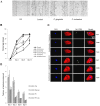
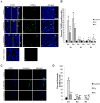
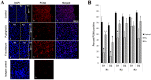
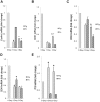
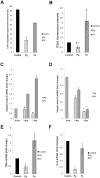
Wounded tissue offers opportunity to microflora to adhere, colonize, invade and infect surrounding healthy tissue. The bacteria of the oral cavity have the potential to alter the wound healing process by interacting with keratinocytes. The aim of this study was to investigate mechanisms through which oral bacteria may influence re-epithelialization by interacting with gingival keratinocytes. By an in vitro scratch assay we demonstrate that primary gingival keratinocytes have impaired closure when exposed to two well characterized oral bacteria, P. gingivalis, and to a lesser extent, F. nucleatum. P. gingivalis reduced wound closure by ∼ 40%, which was partially dependent on proteolytic activity, and bacteria was still present within infected cells 9 days later despite exposure to bacteria for only 24 h. Both oral bacteria caused keratinocyte apoptosis at the wound site with cell death being greatest at the wound edge. P. gingivalis and F. nucleatum adversely affected cell proliferation and the effect also had a spatial component being most striking at the edge. The impact of the bacteria was long lasting even when exposure was brief. Cell migration was compromised in bacteria challenged keratinocytes with P. gingivalis having more severe effect (p<0.05) than F. nucleatum. Quantitative real time PCR of bacteria challenged cells showed that P. gingivalis and to a lesser extent F. nucleatum significantly downregulated cell cycle genes cyclin1, CDK1, and CDK4 (p<0.05) that are critical for GI/S transition. Further, genes associated with cell migration such as integrin beta-3 and -6 were significantly downregulated by P. gingivalis (p<0.05).
Conflict of interest statement
Figures
References
-
- Siddiqui AR, Bernstein JM (2010) Chronic wound infection: facts and controversies. Clin Dermatol 28: 519–526. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

