Oncogenic H-Ras up-regulates acid β-hexosaminidase by a mechanism dependent on the autophagy regulator TFEB
- PMID: 24586816
- PMCID: PMC3933543
- DOI: 10.1371/journal.pone.0089485
Oncogenic H-Ras up-regulates acid β-hexosaminidase by a mechanism dependent on the autophagy regulator TFEB
Abstract
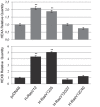
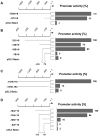
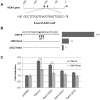
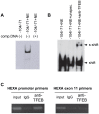
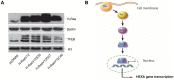
The expression of constitutively active H-RasV12 oncogene has been described to induce proliferative arrest and premature senescence in many cell models. There are a number of studies indicating an association between senescence and lysosomal enzyme alterations, e.g. lysosomal β-galactosidase is the most widely used biomarker to detect senescence in cultured cells and we previously reported that H-RasV12 up-regulates lysosomal glycohydrolases enzymatic activity in human fibroblasts. Here we investigated the molecular mechanisms underlying lysosomal glycohydrolase β-hexosaminidase up-regulation in human fibroblasts expressing the constitutively active H-RasV12. We demonstrated that H-Ras activation increases β-hexosaminidase expression and secretion by a Raf/extracellular signal-regulated protein kinase dependent pathway, through a mechanism that relies on the activity of the transcription factor EB (TFEB). Because of the pivotal role of TFEB in the regulation of lysosomal system biogenesis and function, our results suggest that this could be a general mechanism to enhance lysosomal enzymes activity during oncogene-induced senescence.
Conflict of interest statement
Figures



References
-
- Kornfeld S, Mellman I (1989) The biogenesis of lysosomes. Annu Rev Cell Biol 5: 483–525. - PubMed
-
- Reddy A, Caler E, Andrews NW (2001) Plasma membrane repair is mediated by Ca(2+)-regulated exocytosis of lysosomes. Cell 106: 157–69. - PubMed
-
- Premzl A, Puizdar V, Zavasnik-Bergant V, Kopitar-Jerala N, Lah TT, et al. (2001) Invasion of ras-transformed breast epithelial cells depends on the proteolytic activity of cysteine and aspartic proteinases. Biol Chem 382: 853–7. - PubMed
-
- Urbanelli L, Emiliani C, Massini C, Persichetti E, Orlacchio A, et al. (2008) Cathepsin D expression is decreased in Alzheimer’s disease fibroblasts. Neurobiol Aging 29: 12–22. - PubMed
-
- Fehrenbacher N, Bastholm L, Kirkegaard-Sorensen T, Rafn B, Bottzauw T, et al. (2008) Sensitization to the lysosomal cell death pathway by oncogene-induced down-regulation of lysosome-associated membrane proteins 1 and 2. Cancer Res 68: 6623–33. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

