Assembly of the novel five-component apicomplexan multi-aminoacyl-tRNA synthetase complex is driven by the hybrid scaffold protein Tg-p43
- PMID: 24586818
- PMCID: PMC3930741
- DOI: 10.1371/journal.pone.0089487
Assembly of the novel five-component apicomplexan multi-aminoacyl-tRNA synthetase complex is driven by the hybrid scaffold protein Tg-p43
Abstract
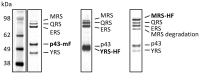
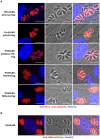
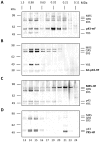
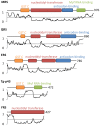
In Toxoplasma gondii, as in other eukaryotes, a subset of the amino-acyl-tRNA synthetases are arranged into an abundant cytoplasmic multi-aminoacyl-tRNA synthetase (MARS) complex. Through a series of genetic pull-down assays, we have identified the enzymes of this complex as: methionyl-, glutaminyl-, glutamyl-, and tyrosyl-tRNA synthetases, and we show that the N-terminal GST-like domain of a partially disordered hybrid scaffold protein, Tg-p43, is sufficient for assembly of the intact complex. Our gel filtration studies revealed significant heterogeneity in the size and composition of isolated MARS complexes. By targeting the tyrosyl-tRNA synthetases subunit, which was found exclusively in the complete 1 MDa complex, we were able to directly visualize MARS particles in the electron microscope. Image analyses of the negative stain data revealed the observed heterogeneity and instability of these complexes to be driven by the intrinsic flexibility of the domain arrangements within the MARS complex. These studies provide unique insights into the assembly of these ubiquitous but poorly understood eukaryotic complexes.
Conflict of interest statement
Figures


References
-
- Szymański M, Deniziak M, Barciszewski J (2000) The new aspects of aminoacyl-tRNA synthetases. Acta Biochim Pol 47: 821–834. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

