A critical evaluation of microRNA biomarkers in non-neoplastic disease
- PMID: 24586876
- PMCID: PMC3935874
- DOI: 10.1371/journal.pone.0089565
A critical evaluation of microRNA biomarkers in non-neoplastic disease
Abstract
Background: MicroRNAs (miRNAs) are small (∼22-nt), stable RNAs that critically modulate post-transcriptional gene regulation. MicroRNAs can be found in the blood as components of serum, plasma and peripheral blood mononuclear cells (PBMCs). Many microRNAs have been reported to be specific biomarkers in a variety of non-neoplastic diseases. To date, no one has globally evaluated these proposed clinical biomarkers for general quality or disease specificity. We hypothesized that the cellular source of circulating microRNAs should correlate with cells involved in specific non-neoplastic disease processes. Appropriate cell expression data would inform on the quality and usefulness of each microRNA as a biomarker for specific diseases. We further hypothesized a useful clinical microRNA biomarker would have specificity to a single disease.
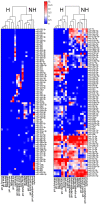
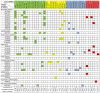
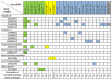
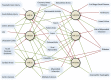
Methods and findings: We identified 416 microRNA biomarkers, of which 192 were unique, in 104 publications covering 57 diseases. One hundred and thirty-nine microRNAs (33%) represented biologically plausible biomarkers, corresponding to non-ubiquitous microRNAs expressed in disease-appropriate cell types. However, at a global level, many of these microRNAs were reported as "specific" biomarkers for two or more unrelated diseases with 6 microRNAs (miR-21, miR-16, miR-146a, miR-155, miR-126 and miR-223) being reported as biomarkers for 9 or more distinct diseases. Other biomarkers corresponded to common patterns of cellular injury, such as the liver-specific microRNA, miR-122, which was elevated in a disparate set of diseases that injure the liver primarily or secondarily including hepatitis B, hepatitis C, sepsis, and myocardial infarction.
Conclusions: Only a subset of reported blood-based microRNA biomarkers have specificity for a particular disease. The remainder of the reported non-neoplastic biomarkers are either biologically implausible, non-specific, or uninterpretable due to limitations of our current understanding of microRNA expression.
Conflict of interest statement
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

