Insight into buffalo (Bubalus bubalis) RIG1 and MDA5 receptors: a comparative study on dsRNA recognition and in-vitro antiviral response
- PMID: 24587036
- PMCID: PMC3935933
- DOI: 10.1371/journal.pone.0089788
Insight into buffalo (Bubalus bubalis) RIG1 and MDA5 receptors: a comparative study on dsRNA recognition and in-vitro antiviral response
Abstract
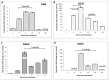
RIG1 and MDA5 have emerged as important intracellular innate pattern recognition receptors that recognize viral RNA and mediate cellular signals controlling Type I interferon (IFN-I) response. Buffalo RIG1 and MDA5 genes were investigated to understand the mechanism of receptor induced antiviral response. Sequence analysis revealed that RIG1 and MDA5 maintain a domain arrangement that is common in mammals. Critical binding site residues of the receptors are evolutionary conserved among mammals. Molecular dynamics simulations suggested that RIG1 and MDA5 follow a similar, if not identical, dsRNA binding pattern that has been previously reported in human. Moreover, binding free energy calculation revealed that MDA5 had a greater affinity towards dsRNA compared to RIG1. Constitutive expressions of RLR genes were ubiquitous in different tissues without being specific to immune organs. Poly I:C stimulation induced elevated expressions of IFN-β and IFN-stimulated genes (ISGs) through interferon regulatory factors (IRFs) mediated pathway in buffalo foetal fibroblast cells. The present study provides crucial insights into the structure and function of RIG1 and MDA5 receptors in buffalo.
Conflict of interest statement
Figures





Similar articles
-
Poly I:C stimulation in-vitro as a marker for an antiviral response in different cell types generated from Buffalo (Bubalus bubalis).Mol Immunol. 2020 May;121:136-143. doi: 10.1016/j.molimm.2020.03.004. Epub 2020 Mar 19. Mol Immunol. 2020. PMID: 32200171
-
Inhibition of antiviral innate immunity by birnavirus VP3 protein via blockage of viral double-stranded RNA binding to the host cytoplasmic RNA detector MDA5.J Virol. 2014 Oct;88(19):11154-65. doi: 10.1128/JVI.01115-14. Epub 2014 Jul 16. J Virol. 2014. PMID: 25031338 Free PMC article.
-
Cooperative assembly and dynamic disassembly of MDA5 filaments for viral dsRNA recognition.Proc Natl Acad Sci U S A. 2011 Dec 27;108(52):21010-5. doi: 10.1073/pnas.1113651108. Epub 2011 Dec 12. Proc Natl Acad Sci U S A. 2011. PMID: 22160685 Free PMC article.
-
LGP2 synergy with MDA5 in RLR-mediated RNA recognition and antiviral signaling.Cytokine. 2015 Aug;74(2):198-206. doi: 10.1016/j.cyto.2015.02.010. Epub 2015 Mar 18. Cytokine. 2015. PMID: 25794939 Free PMC article. Review.
-
MDA5-filament, dynamics and disease.Curr Opin Virol. 2015 Jun;12:20-5. doi: 10.1016/j.coviro.2015.01.011. Epub 2015 Feb 9. Curr Opin Virol. 2015. PMID: 25676875 Free PMC article. Review.
Cited by
-
Comparative genomic analysis of buffalo (Bubalus bubalis) NOD1 and NOD2 receptors and their functional role in in-vitro cellular immune response.PLoS One. 2015 Mar 18;10(3):e0119178. doi: 10.1371/journal.pone.0119178. eCollection 2015. PLoS One. 2015. PMID: 25786158 Free PMC article.
-
Computational Insights into the Structural Dynamics of MDA5 Variants Associated with Aicardi-Goutières Syndrome and Singleton-Merten Syndrome.Biomolecules. 2021 Aug 21;11(8):1251. doi: 10.3390/biom11081251. Biomolecules. 2021. PMID: 34439917 Free PMC article.
-
Preterm birth, intrauterine infection, and fetal inflammation.Front Immunol. 2014 Dec 1;5:574. doi: 10.3389/fimmu.2014.00574. eCollection 2014. Front Immunol. 2014. PMID: 25520716 Free PMC article. Review.
-
Diversity, Antimicrobial Action and Structure-Activity Relationship of Buffalo Cathelicidins.PLoS One. 2015 Dec 16;10(12):e0144741. doi: 10.1371/journal.pone.0144741. eCollection 2015. PLoS One. 2015. PMID: 26675301 Free PMC article.
References
-
- Yoneyama M, Kikuchi M, Natsukawa N, Shinobu N, Imaizumi T, et al. (2004) The RNA helicase RIG1 has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol 5: 730–737. - PubMed
-
- Kawai T, Akira S (2006) Innate immune recognition of viral infection. Nat Immunol 7: 131–137. - PubMed
-
- Sasai M, Shingai M, Funami K, Yoneyama M, Fujita T, et al. (2006) NAK-associated protein 1 participates in both the TLR3 and the cytoplasmic pathways in type I IFN induction. J Immunol 177: 8676–8683. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

