Fast segmentation of stained nuclei in terabyte-scale, time resolved 3D microscopy image stacks
- PMID: 24587204
- PMCID: PMC3937404
- DOI: 10.1371/journal.pone.0090036
Fast segmentation of stained nuclei in terabyte-scale, time resolved 3D microscopy image stacks
Abstract
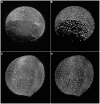
Automated analysis of multi-dimensional microscopy images has become an integral part of modern research in life science. Most available algorithms that provide sufficient segmentation quality, however, are infeasible for a large amount of data due to their high complexity. In this contribution we present a fast parallelized segmentation method that is especially suited for the extraction of stained nuclei from microscopy images, e.g., of developing zebrafish embryos. The idea is to transform the input image based on gradient and normal directions in the proximity of detected seed points such that it can be handled by straightforward global thresholding like Otsu's method. We evaluate the quality of the obtained segmentation results on a set of real and simulated benchmark images in 2D and 3D and show the algorithm's superior performance compared to other state-of-the-art algorithms. We achieve an up to ten-fold decrease in processing times, allowing us to process large data sets while still providing reasonable segmentation results.
Conflict of interest statement
Figures

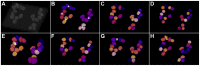
 and
and  (B, C), LoG scale-space maximum intensity projection with
(B, C), LoG scale-space maximum intensity projection with  ,
,  and
and  (D) and the detected seeds plotted on the original image (E).
(D) and the detected seeds plotted on the original image (E).


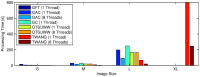
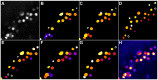
 8000 cells (A,B) and at the 11 hpf stage with
8000 cells (A,B) and at the 11 hpf stage with  12000 cells (C,D), respectively. The panels show maximum intensity projections of the raw images (A,C) and the resulting segmentation using our TWANG segmentation pipeline (B,D). Each 3D image stack has a file size of
12000 cells (C,D), respectively. The panels show maximum intensity projections of the raw images (A,C) and the resulting segmentation using our TWANG segmentation pipeline (B,D). Each 3D image stack has a file size of  GB and comprises 2560×2160×500 voxels with a dynamic range of 16 bits. Processing one image stack takes approximately
GB and comprises 2560×2160×500 voxels with a dynamic range of 16 bits. Processing one image stack takes approximately  minutes on a common desktop machine, depending on the developmental stage of the embryo. Typical experiments may be comprised of up to
minutes on a common desktop machine, depending on the developmental stage of the embryo. Typical experiments may be comprised of up to  z-stacks (
z-stacks ( TB) for the spatio-temporal analysis of a single embryo.
TB) for the spatio-temporal analysis of a single embryo.References
-
- Tomer R, Khairy K, Amat F, Keller P (2012) Quantitative high-speed imaging of entire developing embryos with simultaneous multiview light-sheet microscopy. Nature Methods: 755–763. - PubMed
-
- Huisken J, Swoger J, Del Bene F, Wittbrodt J, Stelzer EH (2004) Optical sectioning deep inside live embryos by selective plane illumination microscopy. Science 305: 1007–1009. - PubMed
-
- Keller P, Schmidt A, Wittbrodt J, Stelzer E (2011) Digital scanned laser light-sheet uorescence microscopy (DSLM) of zebrafish and drosophila embryonic development. Cold Spring Harbor Protocols 2011: pdb–prot065839. - PubMed
-
- de Chaumont F, Dallongeville S, Chenouard N, Hervé N, Pop S, et al. (2012) Icy: An open bioimage informatics platform for extended reproducible research. Nature Methods 9: 690–696. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

