New hydrocarbon degradation pathways in the microbial metagenome from Brazilian petroleum reservoirs
- PMID: 24587220
- PMCID: PMC3935994
- DOI: 10.1371/journal.pone.0090087
New hydrocarbon degradation pathways in the microbial metagenome from Brazilian petroleum reservoirs
Abstract
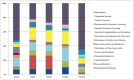
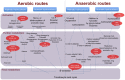
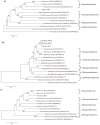
Current knowledge of the microbial diversity and metabolic pathways involved in hydrocarbon degradation in petroleum reservoirs is still limited, mostly due to the difficulty in recovering the complex community from such an extreme environment. Metagenomics is a valuable tool to investigate the genetic and functional diversity of previously uncultured microorganisms in natural environments. Using a function-driven metagenomic approach, we investigated the metabolic abilities of microbial communities in oil reservoirs. Here, we describe novel functional metabolic pathways involved in the biodegradation of aromatic compounds in a metagenomic library obtained from an oil reservoir. Although many of the deduced proteins shared homology with known enzymes of different well-described aerobic and anaerobic catabolic pathways, the metagenomic fragments did not contain the complete clusters known to be involved in hydrocarbon degradation. Instead, the metagenomic fragments comprised genes belonging to different pathways, showing novel gene arrangements. These results reinforce the potential of the metagenomic approach for the identification and elucidation of new genes and pathways in poorly studied environments and contribute to a broader perspective on the hydrocarbon degradation processes in petroleum reservoirs.
Conflict of interest statement
Figures



Similar articles
-
In depth metagenomic analysis in contrasting oil wells reveals syntrophic bacterial and archaeal associations for oil biodegradation in petroleum reservoirs.Sci Total Environ. 2020 May 1;715:136646. doi: 10.1016/j.scitotenv.2020.136646. Epub 2020 Jan 15. Sci Total Environ. 2020. PMID: 32014760
-
Novel, active, and uncultured hydrocarbon-degrading microbes in the ocean.Appl Environ Microbiol. 2024 Sep 18;90(9):e0122424. doi: 10.1128/aem.01224-24. Epub 2024 Aug 23. Appl Environ Microbiol. 2024. PMID: 39177328 Free PMC article.
-
Anaerobic hydrocarbon biodegradation in deep subsurface oil reservoirs.Nature. 2004 Sep 16;431(7006):291-4. doi: 10.1038/nature02922. Nature. 2004. PMID: 15372028
-
Bioremediation of petroleum hydrocarbons: catabolic genes, microbial communities, and applications.Appl Microbiol Biotechnol. 2014 Jun;98(11):4781-94. doi: 10.1007/s00253-014-5684-9. Epub 2014 Apr 2. Appl Microbiol Biotechnol. 2014. PMID: 24691868 Review.
-
Microbial degradation of petroleum hydrocarbons.Bioresour Technol. 2017 Jan;223:277-286. doi: 10.1016/j.biortech.2016.10.037. Epub 2016 Oct 15. Bioresour Technol. 2017. PMID: 27789112 Review.
Cited by
-
Metataxonomic analyses reveal differences in aquifer bacterial community as a function of creosote contamination and its potential for contaminant remediation.Sci Rep. 2019 Aug 13;9(1):11731. doi: 10.1038/s41598-019-47921-y. Sci Rep. 2019. PMID: 31409826 Free PMC article.
-
Carbendazim Modulates the Metabolically Active Bacterial Populations in Soil and Rhizosphere.Curr Microbiol. 2023 Jul 13;80(9):280. doi: 10.1007/s00284-023-03391-0. Curr Microbiol. 2023. PMID: 37439951
-
Genome features of a novel hydrocarbonoclastic Chryseobacterium oranimense strain and its comparison to bacterial oil-degraders and to other C. oranimense strains.DNA Res. 2023 Dec 1;30(6):dsad025. doi: 10.1093/dnares/dsad025. DNA Res. 2023. PMID: 37952165 Free PMC article.
-
Biostimulation of Indigenous Microbial Community for Bioremediation of Petroleum Refinery Sludge.Front Microbiol. 2016 Sep 21;7:1407. doi: 10.3389/fmicb.2016.01407. eCollection 2016. Front Microbiol. 2016. PMID: 27708623 Free PMC article.
-
Culturable hydrocarbonoclastic marine bacterial isolates from Indonesian seawater in the Lombok Strait and Indian Ocean.Heliyon. 2019 May 7;5(5):e01594. doi: 10.1016/j.heliyon.2019.e01594. eCollection 2019 May. Heliyon. 2019. PMID: 31111106 Free PMC article.
References
-
- Torsvik V, Daae FL, Sandaa RA, Ovreas L (1998) Novel techniques for analysing microbial diversity in natural and perturbed environments. Journal of Biotechnology 64: 53–62. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

