Interactions of phosphatase and tensin homologue (PTEN) proteins with phosphatidylinositol phosphates: insights from molecular dynamics simulations of PTEN and voltage sensitive phosphatase
- PMID: 24588644
- PMCID: PMC4167384
- DOI: 10.1021/bi5000299
Interactions of phosphatase and tensin homologue (PTEN) proteins with phosphatidylinositol phosphates: insights from molecular dynamics simulations of PTEN and voltage sensitive phosphatase
Abstract
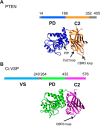
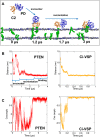
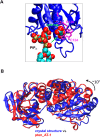
The phosphatase and tensin homologue (PTEN) and the Ciona intestinalis voltage sensitive phosphatase (Ci-VSP) are both phosphatidylinositol phosphate (PIP) phosphatases that contain a C2 domain. PTEN is a tumor suppressor protein that acts as a phosphatase on PIP3 in mammalian cell membranes. It contains two principal domains: a phosphatase domain (PD) and a C2 domain. Despite detailed structural and functional characterization, less is known about its mechanism of interaction with PIP-containing lipid bilayers. Ci-VSP consists of an N-terminal transmembrane voltage sensor domain and a C-terminal PTEN domain, which in turn contains a PD and a C2 domain. The nature of the interaction of the PTEN domain of Ci-VSP with membranes has not been well established. We have used multiscale molecular dynamics simulations to define the interaction mechanisms of PTEN and of the Ci-VSP PTEN domains with PIP-containing lipid bilayers. Our results suggest a novel mechanism of association of the PTEN with such bilayers, in which an initial electrostatics-driven encounter of the protein and bilayer is followed by reorientation of the protein to optimize its interactions with PIP molecules in the membrane. Although a PIP3 molecule binds close to the active site of PTEN, our simulations suggest a further conformational change of the protein may be required for catalytically productive binding to occur. Ci-VSP interacted with membranes in an orientation comparable to that of PTEN but bound directly to PIP-containing membranes without a subsequent reorientation step. Again, PIP3 bound close to the active site of the Ci-VSP PD, but not in a catalytically productive manner. Interactions of Ci-VSP with the bilayer induced clustering of PIP molecules around the protein.
Figures



References
-
- Raimondi C.; Falasca M. (2012) Phosphoinositides signalling in cancer: Focus on PI3K and PLC. Adv. Biol. Regul. 52, 166–182. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
- BB/L002558/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBS/B/16011/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BEP17032/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- B19456/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- 092970/WT_/Wellcome Trust/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

