Quantification of Gram-positive bacteria: adaptation and evaluation of a preparation strategy using high amounts of clinical tissue
- PMID: 24589061
- PMCID: PMC4015715
- DOI: 10.1186/1746-6148-10-53
Quantification of Gram-positive bacteria: adaptation and evaluation of a preparation strategy using high amounts of clinical tissue
Abstract
Background: A preparation method for quantification of bacteria in tissues is obligatory to reduce tissue mass, concentrate the target, purify, remove inhibitory substances and to achieve constant target recovery rates. No preparation method has been available until now for a high mass of tissue applicable for routine use and analytical veterinary diagnostics.
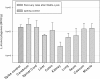
Results: This study describes an easy-to-use tissue preparation protocol to quantify Gram-positive bacteria from a large volume of tissue matrix. A previously published sample preparation method (Matrix-Lysis) from food science was successfully adapted for clinical use on tissues from pigs, including cerebrum, spinal cord, lung, liver, ileum, colon, caecum, kidney and muscle tissue. This tissue preparation method now permits quantification of pathogens from 5 g of organic matrix, which is a 20-200 fold increase by weight compared to other methods. It is based on solubilization of the sample matrix with either a chaotrope plus detergent or divalent salts as solubilization agents. The method was designed as a modular system, offering the possibility to change lysis buffers, according to tissue solubilization characteristics and the intended detection method (molecular or culture). Using Listeria monocytogenes as model organism, viable cell quantification or DNA extraction and quantitative real-time PCR were performed after Matrix-Lysis to determine recovery rates and detection limit (LOD). The adapted Matrix-Lysis protocol resulted in high recovery rates (mean value: 76% ± 39%) for all tested organs, except kidney, and recovery was constant over 5 log scales for all tested buffer systems. The LOD for Matrix-Lysis with subsequent plate count method (PCM) was as low as 1 CFU/5 g, while for qPCR based detection the LOD was 102 bacterial cell equivalents (BCE)/5 g for two buffer systems.
Conclusions: This tissue preparation is inexpensive and can be easily used for routine and analytical veterinary diagnostics. Inoculation studies or hazard assessments can profit from this tissue preparation method and it is anticipated that this study will be a valuable source for further research on tissue preparation strategies.
Figures

References
-
- Espy MJ, Uhl JR, Sloan LM, Buckwalter SP, Jones MF, Vetter EA, Yao JDC, Wengenack NL, Rosenblatt JE, Cockerill FR, Smith TF. Real-time PCR in clinical microbiology: applications for a routine laboratory testing. Clin Microbiol Rev. 2006;19:165–256. doi: 10.1128/CMR.19.1.165-256.2006. - DOI - PMC - PubMed
-
- Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J, Wittwer CT. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55:611–622. doi: 10.1373/clinchem.2008.112797. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

