Hepatitis C virus core protein activates autophagy through EIF2AK3 and ATF6 UPR pathway-mediated MAP1LC3B and ATG12 expression
- PMID: 24589849
- PMCID: PMC5119055
- DOI: 10.4161/auto.27954
Hepatitis C virus core protein activates autophagy through EIF2AK3 and ATF6 UPR pathway-mediated MAP1LC3B and ATG12 expression
Abstract
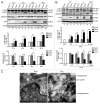
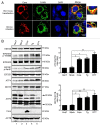
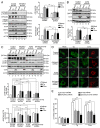
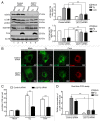
HCV infection induces autophagy, but how this occurs is unclear. Here, we report the induction of autophagy by the structural HCV core protein and subsequent endoplasmic reticular (ER) stress in Huh7 hepatoma cells. During ER stress, both the EIF2AK3 and ATF6 pathways of the unfolded protein response (UPR) were activated by HCV core protein. Then, these pathways upregulated transcription factors ATF4 and DDIT3. The ERN1-XBP1 pathway was not activated. Through ATF4 in the EIF2AK3 pathway, the autophagy gene ATG12 was upregulated. DDIT3 upregulated the transcription of autophagy gene MAP1LC3B (LC3B) by directly binding to the -253 to -99 base region of the LC3B promoter, contributing to the development of autophagy. Collectively, these data suggest not only a novel role for the HCV core protein in autophagy but also offer new insight into detailed molecular mechanisms with respect to HCV-induced autophagy, specifically how downstream UPR molecules regulate key autophagic gene expression.
Keywords: ER stress; UPR; autophagy; core; hepatitis C virus.
Figures






Similar articles
-
Silica nanoparticles induce autophagosome accumulation via activation of the EIF2AK3 and ATF6 UPR pathways in hepatocytes.Autophagy. 2018;14(7):1185-1200. doi: 10.1080/15548627.2018.1458174. Epub 2018 Jul 20. Autophagy. 2018. PMID: 29940794 Free PMC article.
-
MTORC1 coordinates the autophagy and apoptosis signaling in articular chondrocytes in osteoarthritic temporomandibular joint.Autophagy. 2020 Feb;16(2):271-288. doi: 10.1080/15548627.2019.1606647. Epub 2019 Apr 21. Autophagy. 2020. PMID: 31007149 Free PMC article.
-
The unfolded protein response protects human tumor cells during hypoxia through regulation of the autophagy genes MAP1LC3B and ATG5.J Clin Invest. 2010 Jan;120(1):127-41. doi: 10.1172/JCI40027. Epub 2009 Dec 14. J Clin Invest. 2010. PMID: 20038797 Free PMC article.
-
ER Stress and Unfolded Protein Response in Leukemia: Friend, Foe, or Both?Biomolecules. 2021 Jan 30;11(2):199. doi: 10.3390/biom11020199. Biomolecules. 2021. PMID: 33573353 Free PMC article. Review.
-
[Pathogenic Mechanism of Diabetes Development Due to Dysfunction of Unfolded Protein Response].Yakugaku Zasshi. 2016;136(6):817-25. doi: 10.1248/yakushi.15-00292-4. Yakugaku Zasshi. 2016. PMID: 27252061 Review. Japanese.
Cited by
-
Melatonin inhibits bovine viral diarrhea virus replication by ER stress-mediated NF-κB signal pathway and autophagy in MDBK cells.Front Cell Infect Microbiol. 2024 Aug 21;14:1431836. doi: 10.3389/fcimb.2024.1431836. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 39233905 Free PMC article.
-
The Multifaceted Roles of Autophagy in Flavivirus-Host Interactions.Int J Mol Sci. 2018 Dec 7;19(12):3940. doi: 10.3390/ijms19123940. Int J Mol Sci. 2018. PMID: 30544615 Free PMC article. Review.
-
Effects of Spatial Expression of Activating Transcription Factor 4 on the Pathogenicity of Two Phenotypes of Bovine Viral Diarrhea Virus by Regulating the Endoplasmic Reticulum-Mediated Autophagy Process.Microbiol Spectr. 2023 Mar 20;11(2):e0422522. doi: 10.1128/spectrum.04225-22. Online ahead of print. Microbiol Spectr. 2023. PMID: 36939351 Free PMC article.
-
Sirtuin 6 overexpression relieves sepsis-induced acute kidney injury by promoting autophagy.Cell Cycle. 2019 Feb;18(4):425-436. doi: 10.1080/15384101.2019.1568746. Epub 2019 Jan 30. Cell Cycle. 2019. PMID: 30700227 Free PMC article.
-
ATF6-Mediated Unfolded Protein Response Facilitates Adeno-associated Virus 2 (AAV2) Transduction by Releasing the Suppression of the AAV Receptor on Endoplasmic Reticulum Stress.J Virol. 2022 Feb 9;96(3):e0110321. doi: 10.1128/JVI.01103-21. Epub 2021 Dec 1. J Virol. 2022. PMID: 34851146 Free PMC article.
References
-
- Ray Kim W. . Global epidemiology and burden of hepatitis C. Microbes Infect 2002; 4:1219 - 25; http://dx.doi.org/10.1016/S1286-4579(02)01649-0; PMID: 12467763 - DOI - PubMed
-
- Moradpour D, Penin F, Rice CM. . Replication of hepatitis C virus. Nat Rev Microbiol 2007; 5:453 - 63; http://dx.doi.org/10.1038/nrmicro1645; PMID: 17487147 - DOI - PubMed
-
- Gale M Jr., Foy EM. . Evasion of intracellular host defence by hepatitis C virus. Nature 2005; 436:939 - 45; http://dx.doi.org/10.1038/nature04078; PMID: 16107833 - DOI - PubMed
-
- Egger D, Wölk B, Gosert R, Bianchi L, Blum HE, Moradpour D, Bienz K. . Expression of hepatitis C virus proteins induces distinct membrane alterations including a candidate viral replication complex. J Virol 2002; 76:5974 - 84; http://dx.doi.org/10.1128/JVI.76.12.5974-5984.2002; PMID: 12021330 - DOI - PMC - PubMed
-
- Gosert R, Egger D, Lohmann V, Bartenschlager R, Blum HE, Bienz K, Moradpour D. . Identification of the hepatitis C virus RNA replication complex in Huh-7 cells harboring subgenomic replicons. J Virol 2003; 77:5487 - 92; http://dx.doi.org/10.1128/JVI.77.9.5487-5492.2003; PMID: 12692249 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
