Evolution of transcription factor binding in metazoans - mechanisms and functional implications
- PMID: 24590227
- PMCID: PMC4175440
- DOI: 10.1038/nrg3481
Evolution of transcription factor binding in metazoans - mechanisms and functional implications
Abstract
Differences in transcription factor binding can contribute to organismal evolution by altering downstream gene expression programmes. Genome-wide studies in Drosophila melanogaster and mammals have revealed common quantitative and combinatorial properties of in vivo DNA binding, as well as marked differences in the rate and mechanisms of evolution of transcription factor binding in metazoans. Here, we review the recently discovered rapid 're-wiring' of in vivo transcription factor binding between related metazoan species and summarize general principles underlying the observed patterns of evolution. We then consider what might explain the differences in genome evolution between metazoan phyla and outline the conceptual and technological challenges facing this research field.
Figures


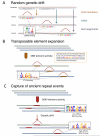
References
-
- Arendt D. The evolution of cell types in animals: emerging principles from molecular studies. Nat Rev Genet. 2008;9:868–82. - PubMed
-
- Shubin N, Tabin C, Carroll S. Deep homology and the origins of evolutionary novelty. Nature. 2009;457:818–23. - PubMed
-
- Brawand D, et al. The evolution of gene expression levels in mammalian organs. Nature. 2011;478:343–8. - PubMed
-
- Vaquerizas JM, Kummerfeld SK, Teichmann SA, Luscombe NM. A census of human transcription factors: function, expression and evolution. Nat Rev Genet. 2009;10:252–63. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

