Fibrotic extracellular matrix activates a profibrotic positive feedback loop
- PMID: 24590289
- PMCID: PMC3971953
- DOI: 10.1172/JCI71386
Fibrotic extracellular matrix activates a profibrotic positive feedback loop
Abstract
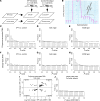
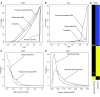
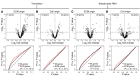
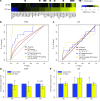
Pathological remodeling of the extracellular matrix (ECM) by fibroblasts leads to organ failure. Development of idiopathic pulmonary fibrosis (IPF) is characterized by a progressive fibrotic scarring in the lung that ultimately leads to asphyxiation; however, the cascade of events that promote IPF are not well defined. Here, we examined how the interplay between the ECM and fibroblasts affects both the transcriptome and translatome by culturing primary fibroblasts generated from IPF patient lung tissue or nonfibrotic lung tissue on decellularized lung ECM from either IPF or control patients. Surprisingly, the origin of the ECM had a greater impact on gene expression than did cell origin, and differences in translational control were more prominent than alterations in transcriptional regulation. Strikingly, genes that were translationally activated by IPF-derived ECM were enriched for those encoding ECM proteins detected in IPF tissue. We determined that genes encoding IPF-associated ECM proteins are targets for miR-29, which was downregulated in fibroblasts grown on IPF-derived ECM, and baseline expression of ECM targets could be restored by overexpression of miR-29. Our data support a model in which fibroblasts are activated to pathologically remodel the ECM in IPF via a positive feedback loop between fibroblasts and aberrant ECM. Interrupting this loop may be a strategy for IPF treatment.
Figures


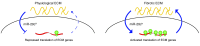
References
-
- Schor SL. Cell proliferation and migration on collagen substrata in vitro. J Cell Sci. 1980;41(1):159–175. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

