Mechanism of Escherichia coli resistance to Pyrrhocoricin
- PMID: 24590485
- PMCID: PMC3993218
- DOI: 10.1128/AAC.02565-13
Mechanism of Escherichia coli resistance to Pyrrhocoricin
Abstract
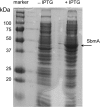
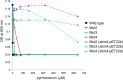
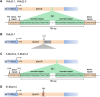
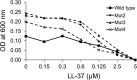
Due to their lack of toxicity to mammalian cells and good serum stability, proline-rich antimicrobial peptides (PR-AMPs) have been proposed as promising candidates for the treatment of infections caused by antimicrobial-resistant bacterial pathogens. It has been hypothesized that these peptides act on multiple targets within bacterial cells, and therefore the likelihood of the emergence of resistance was considered to be low. Here, we show that spontaneous Escherichia coli mutants resistant to pyrrhocoricin arise at a frequency of approximately 6 × 10(-7). Multiple independently derived mutants all contained a deletion in a nonessential gene that encodes the putative peptide uptake permease SbmA. Sensitivity could be restored to the mutants by complementation with an intact copy of the sbmA gene. These findings question the viability of the development of insect PR-AMPs as antimicrobials.
Figures



References
-
- Centers for Disease Control and Prevention (CDC). 2013. Antibiotic resistance threats in the United States, 2013. CDC, Atlanta, GA: http://www.cdc.gov/drugresistance/threat-report-2013
-
- Hancock REW. 2007. The end of an era. Nat. Rev. Drug Discov. 6:28. 10.1038/nrd2223 - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

