Interplay of host microbiota, genetic perturbations, and inflammation promotes local development of intestinal neoplasms in mice
- PMID: 24590763
- PMCID: PMC3949565
- DOI: 10.1084/jem.20131587
Interplay of host microbiota, genetic perturbations, and inflammation promotes local development of intestinal neoplasms in mice
Abstract
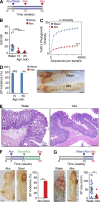
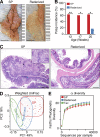
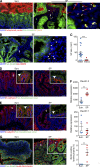
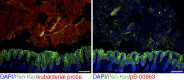
The preferential localization of some neoplasms, such as serrated polyps (SPs), in specific areas of the intestine suggests that nongenetic factors may be important for their development. To test this hypothesis, we took advantage of transgenic mice that expressed HB-EGF throughout the intestine but developed SPs only in the cecum. Here we show that a host-specific microbiome was associated with SPs and that alterations of the microbiota induced by antibiotic treatment or by embryo transfer rederivation markedly inhibited the formation of SPs in the cecum. Mechanistically, development of SPs was associated with a local decrease in epithelial barrier function, bacterial invasion, production of antimicrobials, and increased expression of several inflammatory factors such as IL-17, Cxcl2, Tnf-α, and IL-1. Increased numbers of neutrophils were found within the SPs, and their depletion significantly reduced polyp growth. Together these results indicate that nongenetic factors contribute to the development of SPs and suggest that the development of these intestinal neoplasms in the cecum is driven by the interplay between genetic changes in the host, an inflammatory response, and a host-specific microbiota.
Figures



Comment in
-
Do bugs define cancer geography?J Exp Med. 2014 Mar 10;211(3):385. doi: 10.1084/jem.2113insight3. J Exp Med. 2014. PMID: 24616475 Free PMC article. No abstract available.
References
-
- Bindea G., Mlecnik B., Hackl H., Charoentong P., Tosolini M., Kirilovsky A., Fridman W.H., Pagès F., Trajanoski Z., Galon J. 2009. ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 25:1091–1093 10.1093/bioinformatics/btp101 - DOI - PMC - PubMed
-
- Bongers G., Maussang D., Muniz L.R., Noriega V.M., Fraile-Ramos A., Barker N., Marchesi F., Thirunarayanan N., Vischer H.F., Qin L., et al. 2010. The cytomegalovirus-encoded chemokine receptor US28 promotes intestinal neoplasia in transgenic mice. J. Clin. Invest. 120:3969–3978 10.1172/JCI42563 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

