Surface dynamics of GluN2B-NMDA receptors controls plasticity of maturing glutamate synapses
- PMID: 24591565
- PMCID: PMC4194110
- DOI: 10.1002/embj.201386356
Surface dynamics of GluN2B-NMDA receptors controls plasticity of maturing glutamate synapses
Abstract
NMDA-type glutamate receptors (NMDAR) are central actors in the plasticity of excitatory synapses. During adaptive processes, the number and composition of synaptic NMDAR can be rapidly modified, as in neonatal hippocampal synapses where a switch from predominant GluN2B- to GluN2A-containing receptors is observed after the induction of long-term potentiation (LTP). However, the cellular pathways by which surface NMDAR subtypes are dynamically regulated during activity-dependent synaptic adaptations remain poorly understood. Using a combination of high-resolution single nanoparticle imaging and electrophysiology, we show here that GluN2B-NMDAR are dynamically redistributed away from glutamate synapses through increased lateral diffusion during LTP in immature neurons. Strikingly, preventing this activity-dependent GluN2B-NMDAR surface redistribution through cross-linking, either with commercial or with autoimmune anti-NMDA antibodies from patient with neuropsychiatric symptoms, affects the dynamics and spine accumulation of CaMKII and impairs LTP. Interestingly, the same impairments are observed when expressing a mutant of GluN2B-NMDAR unable to bind CaMKII. We thus uncover a non-canonical mechanism by which GluN2B-NMDAR surface dynamics plays a critical role in the plasticity of maturing synapses through a direct interplay with CaMKII.
Figures

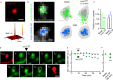
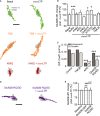
Upper panels: Representative dendritic fragments of a hippocampal neuron (13 div) expressing GluA1-SEP and Homer 1c-DsRed. Clusters of GluA1-SEP were imaged for at least 30 min. The pseudocolor representation shows the different intensity levels of the GluA1-SEP staining. GluA1-SEP intensity was measured in synapses (Homer1c cluster) before and after the induction of chemLTP (see Materials and Methods). Note that 10 min after chemLTP, the GluA1-SEP fluorescence intensity increased in postsynaptic clusters. Arrows designate synapses (i.e., Homer). Bottom panels: magnifications and time-lapses of the outlined GluA1-SEP synaptic clusters. Scale bar = 5 μm; insets = 500 nm; bottom panels = 350 nm.
Left panel: Representative example of a GluA1-SEP fluorescence time course before and after chemLTP (red) or buffer (black) protocol. The average GluA1-SEP synaptic intensity was significantly increased after chemLTP (24 min after induction) (middle panel). Student's t-test, ***P < 0.001. Right panel: distribution of the change in synaptic GluA1-SEP content under control (black line, n = 364 syn. clusters) and chemLTP (red line, n = 837) conditions. Note the right shift in the distribution, indicating an increased proportion of synaptic GluA1-AMPAR after chemLTP.
Left: Schematic drawing of a single QD–antibody complex targeting a surface NMDAR. Right: Representative trajectories (25-s duration, 30-Hz acquisition) of surface QD-conjugated GluN2B- (top, green line) and GluN2A-NMDAR (bottom, blue line) in Homer 1c-GFP-labeled synaptic areas (black dotted line) in an immature hippocampal neuron (11 div). Scale bar = 5 μm.
Representative GluN2A- and GluN2B-NMDAR trajectories during baseline and right after chemLTP (1–4 min). Note the lateral displacement of synaptic GluN2B-NMDAR after chemLTP. The postsynaptic densities are represented by the gray shapes. Scale bar = 0.75 μm.
Comparison of the instantaneous membrane diffusion coefficients of synaptic GluN2A- and GluN2B-NMDAR in different experimental conditions. Note that GluN2B-NMDAR membrane diffusion coefficients are significantly increased right after chemLTP (1–2 min; n = 15, *P < 0.05). This increase is prevented by a bath application of a competitive NMDAR antagonist, AP5 (50 μM, n = 6, Mann-Whitney test, P > 0.05). GluN2A-NMDAR membrane diffusion coefficients are not significantly changed by chemLTP (n = 19, P > 0.05). The symbols represent the mean ± s.e.m.
Comparison of the chemLTP-elicited change in synaptic GluN2B-NMDAR surface diffusion (median normalized to baseline for each condition) in immature (7–12 div) and mature (14–21 div) neurons. Bar graphs represent mean ± s.e.m. Mann-Whitney test, *P < 0.05.
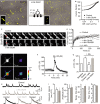
Detection of a single QD (30-Hz acquisition) in our experimental conditions. The high signal-to-noise ratio (SNR) (> 5) enables the detection and location of the signal with a high pointing accuracy (˜30 nm). The QD fluorescence is quantified on a pseudocolor scale (low: red; high: yellow). Scale bar = 800 nm.
A 500-frame stack is obtained while tracking down a single NMDAR/QD complex. On each frame, a single GluN2B- (green) or GluN2A- (blue) QD particle complex is detected and precisely located within synaptic (dark gray) and perisynaptic (320-nm annulus around the synapse; light gray) areas. Those 500 locations are then projected on a single image, providing the successive positions of this receptor/particle complex during the 500-frame stack. Note that GluN2A-NMDAR are more concentrated within the core of the PSD. Scale bar image = 300 nm; synaptic areas = 200 nm.
Relative fraction of synaptic and perisynaptic GluN2-QD particles, calculated before and after chemLTP for both GluN2B- (left) and GluN2A-NMDAR (right) (n = 25 and 20 dendritic fields before and after chemLTP, respectively). Note the significant decrease in the relative synaptic content in GluN2B-NMDAR particles right after chemLTP (Student's t-test, **P < 0.01).
Time-lapse imaging of GluN2B-SEP in an immature hippocampal neuron (10 div) before and after chemLTP. The GluN2B-SEP fluorescence was followed in glutamate postsynaptic area (Homer 1c-DsRed; white dotted line). Homer 1c fluorescence was imaged at the beginning and at the end of the recording (35 min). Note that the Homer 1c fluorescence intensity and area increased 35 min after chemLTP (yellow full line). Scale bar = 800 nm.
Representative time-lapse recording of GluN2B-SEP fluorescence within synapses (Homer 1c area) before and after buffer (gray dots) and chemLTP (green dots) application.
Average GluN2B-SEP fluorescence before (basal) and 30 min after buffer (gray dot; n = 14; paired test, P > 0.05) or chemLTP (green dot; n = 23; paired Student's t-test, **P < 0.01).
Trajectories of single surface QD-GluN1-NMDAR (30-Hz acquisition, 30-s duration) in hippocampal neurons in control (left) and GluN1x-link (right) conditions. A schematic representation of the NMDAR x-link technique using primary anti-GluN (Iary Ab) and secondary (IIary Ab) antibodies is shown in the middle panel. Insets: enlarged GluN1-QD trajectories. Field scale bar = 5 μm; inset scale bar = 1 μm.
Cumulative distribution of GluN1-NMDAR instantaneous surface diffusion coefficients in control and GluN1x-link conditions. Note the leftward shift of the curve in the GluN1x-link condition, indicating a slowdown of surface diffusion.
Representative FRAP acquisition of GluA1-SEP in control and GluN1-NMDAR x-link conditions. The circles indicate the bleached regions. Scale bar = 5 μm.
Average FRAP recovery curves of GluA1-SEP fluorescence in control (n = 13), GluN1 x-link (n = 3), and GluN2B x-link (n = 4) conditions. Full lines represent the average recovery, while dotted lines represent the mean ± s.e.m. The fluorescence recovery of surface synaptic GluA1-SEP remained unaffected in all conditions (P > 0.05).
Representative images of a hippocampal neuron in the basal condition or after glutamate (30 μM) application. The pseudocolor representation shows the different intensity levels of the calcium indicator (Fluo4-AM, 2 μM) before and after the glutamate stimulation. Scale bar = 20 μm.
Average calcium intensity change (ΔF/F0) over time after glutamate stimulation of hippocampal neurons (n = 29).
Relative comparison (percent of basal) of a transient calcium rise induced by glutamate in control (n = 29), control + AP5 (n = 12, Student's t-test, **P < 0.01 when compared to control), and GluN1-NMDAR x-link (n = 21).
Representative recordings of spontaneous NMDAR-mediated EPSC in control (black trace) or GluN1 x-link (gray trace) condition. Averaged sEPSC are shown below. Note the perfect overlay of the averaged NMDAR sEPSC in both conditions.
Comparisons of NMDAR sEPSC inter-event interval, amplitude, decay, and rise times (control, n = 8; GluN1 x-link, n = 9; Student's t-test, P > 0.05 for all parameters).
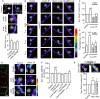
Comparison of the GluA1-SEP fluorescence within synapses (white dotted circle) in spines from control (n = 786 synapses), GluN1 x-link (n = 1324), or GluN2B x-link (n = 987) condition. The dendritic shaft is indicated by the arrow head. Scale bar = 1 μm. The bar graphs represent the mean value ± s.e.m.
Time-lapse imaging of spine areas containing GluA1-SEP (white dotted circle) from immature hippocampal neurons in control (no stimulation), chemLTP, chemLTP + GluN1 x-link, and chemLTP + GluN2B x-link conditions. The pseudocolor representation shows the different intensity levels of the GluA1-SEP staining. Scale bar = 1 μm.
Comparison of the normalized GluA1-SEP fluorescence intensity within synapses in control (n = 786 synapses), chemLTP alone (n = 1324, ***P < 0.001 when compared to control), chemLTP + GluN1 x-link (n = 736, P > 0.05), and chemLTP + GluN2B x-link (n = 966, P > 0.05) conditions. ANOVA followed by Newman-Keuls Multiple Comparison Test.
Comparison of the percentage of potentiated synapses in control (n = 364), chemLTP alone (n = 837, ***P < 0.001), chemLTP + GluN1 x-link (n = 312, P > 0.05), and chemLTP + GluN2B x-link (n = 472 syn. clusters, P > 0.05) conditions. ANOVA followed by Newman-Keuls Multiple Comparison Test.
Representative image of surface GluA2-AMPAR (green) and postsynaptic density protein Shank (red) immunostaining. Scale bar = 5 μm. Right panel, GluA2-SEP fluorescence was detected in spines in control, GluN1 x-link, and GluN2B x-link, before and after chemLTP. Scale bar = 500 nm.
Quantification of GluA2-AMPAR content in synapses. As for GluA1-AMPAR, chemLTP (n = 98 dendritic fields) increased GluA2-AMPAR synaptic content. This was blocked in GluN1- (n = 24 dendritic fields) and GluN2B-NMDAR (n = 6 dendritic fields) x-link conditions when compared to their respective control conditions (n = 113, n = 19 and n = 12 dendritic fields, respectively). ANOVA followed by Newman-Keuls Multiple Comparison Test, ***P < 0.001.
Bar graph: Comparison of the GluA1-SEP fluorescence content within synapses in control (n = 370 synapses), forskolin/rolipram (FSK/Rol.; n = 550 synapses; *P < 0.05), and FSK/Rol. + GluN1 x-link (n = 558 synapses; *P < 0.05) conditions. ANOVA followed by Newman-Keuls Multiple Comparison Test. Data are presented as mean ± s.e.m. Top panels: representative images showing increased GluA1-SEP fluorescence within synapses (white dotted circles) after forskolin/rolipram application. Scale bar = 2.5 μm.

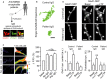
Schematic diagram of the anti-NMDAR IgG isolation procedure from anti-NMDAR encephalitis patients. The cerebrospinal fluid (CSF) was collected and IgG were purified for in vitro imaging experiments. Lower panels: note the high co-localization of surface staining from surface patient anti-NMDAR IgG (“sPat. IgG,” green) and commercial anti-GluN1 antibodies (“sGluN1,” red). Scale bar = 1 μm.
Representative GluN2B-NMDAR-QD trajectories from neurons incubated either with control or with patient IgG. Note the massive reduction in surface dynamics. Scale bar = 250 nm.
Representative images of hippocampal neurons in the basal conditions or after glutamate (30 μM) application. The pseudocolor representation shows the different intensity levels of the calcium indicator (Fluo4-AM, 2 μM) before and after the glutamate stimulation. Neurons were incubated either with no IgG, controls' IgG (Cont. IgG), or patients' IgG (Pat. IgG). Scale bar = 20 μm. Right panel: Average calcium intensity change (ΔF/F0) over time after glutamate stimulation of hippocampal neurons in no IgG, controls' IgG (Cont. IgG), or patients' IgG (Pat. IgG) conditions.
Hippocampal neurons expressing either GluN1-SEP or GluA1-SEP were incubated with IgG (5 μg/ml) either from control or from anti-NMDAR patients for 20–25 min. Note that patient IgG do not affect GluN1-SEP distribution. Neurons were stimulated with a chemLTP protocol and each synaptic GluA1-AMPAR cluster was followed over time. Note that chemLTP increased the intensity of GluA1-SEP in synaptic clusters (arrows) only in control IgG condition. Scale bars = 1 μm. Lower panels: Quantification of the GluA1-AMPAR synaptic content and percentage of potentiated GluA1-AMPAR synapses in control or patient IgG conditions. For each neuron, GluA1 synaptic fluorescence intensity was quantified before and 10–15 min after chemLTP. The GluA1-AMPAR synaptic content and percentage of potentiated GluA1-AMPAR synapses significantly increased in control condition (n = 6 neurons; Student's t-test, *P < 0.05 for GluA1 synaptic content, **P < 0.01 for the percentage of potentiated GluA1 synapses) but not in the presence of patient IgG (n = 9, P > 0.05 in all parameters). ANOVA followed by Newman-Keuls Multiple Comparison Test. Bars represent mean ± s.e.m.
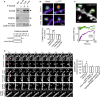
Representative trajectories (25-s duration, 20-Hz acquisition) of surface QD-conjugated GluN2B-NMDAR within Homer 1c-GFP-labeled synaptic areas (gray) before and after chemLTP in immature hippocampal neuron (9–12 div) in control (green), TBB (orange), and KN62 (red) conditions. Scale bar = 1 μm.
Synaptic GluN2B-NMDAR surface diffusion change (normalized to baseline for each condition) after chemLTP in immature neurons (< 12 div) pre-incubated (15 min) with either buffer (PBS, n = 3,388 trajectories, ***P < 0.001), CaMKII inhibitors KN93 (5 μM, n = 1,521, P > 0.05), AIP2 (5 μM, n = 1,199, P > 0.05), or KN62 (5 μM, n = 3,088, P > 0.05), PKC inhibitor Gö6976 (10 μM, n = 3,175, *P < 0.05), PKA inhibitor KT5720 (10 μM, n = 847, *P < 0.05), and CK2 inhibitors TBB (10 μM, 4 h pre-incubation, n = 3,322, P > 0.05) or TMCB (10 μM, n = 1,200, P > 0.05). Kruskal-Wallis followed by Dunn‘s Multiple Comparison Test, for all P-values.
Mean extrasynaptic and synaptic GluN2B-NMDAR diffusion coefficients in control condition (extrasynaptic, n = 6,160 trajectories; synaptic, n = 4,199 trajectories), in the presence of the CK2 inhibitor TMCB (10 μM; extrasynaptic, n = 7,889 trajectories; synaptic, n = 2,427 trajectories; Mann-Whitney test, P > 0.05 compared to control) or the CaMKII inhibitor AIP2 (5 μM; extrasynaptic, n = 772 trajectories; synaptic, n = 86 trajectories; Mann-Whitney test, ***P < 0.0001 compared to control), and mean extrasynaptic and synaptic GluN2B-(R1300Q/S1303D)-NMDAR diffusion coefficients (GluN2B-RQ/SD; extrasynaptic, n = 880 trajectories; synaptic, n = 29 trajectories; Mann-Whitney test, ***P < 0.0001 compared to control).
Representative trajectories (25-s duration, 20-Hz acquisition) of surface QD-conjugated GluN2B-(R1300Q/S1303D)-NMDAR(GluN2B-RQ/SD) within Homer 1c-GFP-labeled synaptic areas (gray) before and after chemLTP in immature hippocampal neuron (9–12 div). Scale bar = 1 μm.
Synaptic GluN2B-SEP and GluN2B-RQ/SD surface diffusion change (normalized to baseline for each condition) after chemLTP (GluN2B-SEP, n = 1,498 trajectories, baseline versus chemLTP, Student's t-test, **P < 0.01; GluN2B-RQ/SD, n = 323 trajectories, baseline versus chemLTP, Mann-Whitney test, P < 0.05).
Representative immunoblots showing the immunoprecipitation (IP) of CaMKII (α form) and phospho-CaMKII-Thr286 with GluN2B in membrane fractions from hippocampal slices (P17–20 rats) incubated with buffer (control) or GluN1 x-link. Lower panel: the ratio between CaMKII and GluN2B optical densities is represented (n = 3 independent experiments). SM, start material; No Ab., no antibody; Cont., control.
CaMKII-GFP was detected and imaged in spines before (basal) and after chemLTP in control and GluN1x-link conditions. Scale bar = 1 μm. Lower panel: CaMKII-GFP fluorescence intensity was compared between these conditions (basal: n = 6 neuronal fields, N = 765 spines; chemLTP: n = 11 neuronal fields, N = 1,688 spines; basal versus chemLTP, Student's t-test, ***P < 0.0001; x-link GluN1: n = 7 neuronal fields, N = 955 spines; x-link GluN1+chemLTP: n = 9 neuronal fields, N = 1,728 spines; x-link GluN1 versus x-link GluN1+chemLTP, Student's t-test, *P < 0.05).
Time-lapse imaging of CaMKII-GFP in hippocampal dendritic spines in basal (light and dark green dotted circles) or FRAP (purple dotted circle) conditions. Scale bar = 1 μm. Lower panel: the fluorescence intensity was measured and plotted over time. CaMKII-GFP fluorescence was stable over time in basal condition. Note the recovery after photobleaching of the CaMKII-GFP fluorescence up to nearly 60% of its initial value, indicating a 60% mobile fraction.
FRAP imaging of CaMKII-GFP in control, chemLTP, GluN1 x-link, GluN1 x-link +chemLTP, GluN2B-RQ/SD, and GluN2B-RQ/SD + chemLTP conditions. Scale bar = 2 μm. The dotted line circles represent the photobleached areas. Time (t) is expressed in seconds. Right panel: normalized variation of the CaMKII-GFP mobile fraction in basal (n = 6 dendritic fields), chemLTP (n = 10 dendritic fields, Student's t-test, *P < 0.05 compared to control), GluN1 x-link (black, n = 6 dendritic fields), GluN1 x-link+chemLTP (n = 11 dendritic fields, Student's t-test, P > 0.05 when compared to GluN1 x-link), GluN2B-RQ/SD (n = 9 dendritic fields), and GluN2B-RQ/SD+chemLTP (Student t-test n = 19 dendritic fields, Student's t-test, P > 0.05 when compared to GluN2B-RQ/SD) conditions.

Comment in
-
LTP: GluN2B on the go.EMBO J. 2014 Apr 16;33(8):781-2. doi: 10.1002/embj.201487958. Epub 2014 Mar 5. EMBO J. 2014. PMID: 24599308 Free PMC article.
References
-
- Abrahamsson T, Gustafsson B, Hanse E. AMPA silencing is a prerequisite for developmental long-term potentiation in the hippocampal CA1 region. J Neurophysiol. 2008;100:2605–2614. - PubMed
-
- Barria A, Malinow R. NMDA receptor subunit composition controls synaptic plasticity by regulating binding to CaMKII. Neuron. 2005;48:289–301. - PubMed
-
- Bashir ZI, Alford S, Davies SN, Randall AD, Collingridge GL. Long-term potentiation of NMDA receptor-mediated synaptic transmission in the hippocampus. Nature. 1991;349:156–158. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

