Strength of a bifurcated H bond
- PMID: 24591597
- PMCID: PMC3964065
- DOI: 10.1073/pnas.1319827111
Strength of a bifurcated H bond
Abstract
Macromolecules are characterized by their particular arrangement of H bonds. Many of these interactions involve a single donor and acceptor pair, such as the regular H-bonding pattern between carbonyl oxygens and amide H(+)s four residues apart in α-helices. The H-bonding potential of some acceptors, however, leads to the phenomenon of overcoordination between two donors and one acceptor. Herein, using isotope-edited Fourier transform infrared measurements and density functional theory (DFT) calculations, we measured the strength of such bifurcated H bonds in a transmembrane α-helix. Frequency shifts of the (13)C=(18)O amide I mode were used as a reporter of the strength of the bifurcated H bond from a thiol and hydroxyl H(+) at residue i + 4. DFT calculations yielded very similar frequency shifts and an energy of -2.6 and -3.4 kcal/mol for the thiol and hydroxyl bifurcated H bonds, respectively. The strength of the intrahelical bifurcated H bond is consistent with its prevalence in hydrophobic environments and is shown to significantly impact side-chain rotamer distribution.
Keywords: FTIR; membrane proteins; protein structure.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
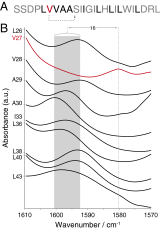
 C=
C= O. Val27 is in red because it is the only residue in the sequence that has a hydroxylic residue four residues in its C-terminal direction, as indicated by the dotted arrow. Note that numbering is according to the sequence of the full length protein. (B) FTIR spectra in the region of the isotope-edited amide I peak of 10 M2 transmembrane peptides in hydrated lipid bilayers (35). The different peptides are labeled with a 1-
O. Val27 is in red because it is the only residue in the sequence that has a hydroxylic residue four residues in its C-terminal direction, as indicated by the dotted arrow. Note that numbering is according to the sequence of the full length protein. (B) FTIR spectra in the region of the isotope-edited amide I peak of 10 M2 transmembrane peptides in hydrated lipid bilayers (35). The different peptides are labeled with a 1- C=
C= O at the position indicated on the ordinate. The shaded region represents the peak center range for all peptides except for the peptide labeled at Val27. The wavenumber shift (cm
O at the position indicated on the ordinate. The shaded region represents the peak center range for all peptides except for the peptide labeled at Val27. The wavenumber shift (cm ) between the Val27 peak and the average of all of the other peptides is indicated.
) between the Val27 peak and the average of all of the other peptides is indicated.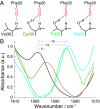
 C and
C and  O) labeled carbonyl group of Phe26 is depicted in red. The main-chain (canonical) and bifurcated H bonds are shown in orange and purple, respectively. (B) FTIR spectra in the region of the isotope-edited amide I peak of four SARS E peptides in hydrated lipid bilayers. The different peptides are: Val30 (black), Cys30 (brown), Thr30 (green), and Ser30 (cyan). The wavenumber shifts relative to the peptide with a valine at position 30 are indicated.
O) labeled carbonyl group of Phe26 is depicted in red. The main-chain (canonical) and bifurcated H bonds are shown in orange and purple, respectively. (B) FTIR spectra in the region of the isotope-edited amide I peak of four SARS E peptides in hydrated lipid bilayers. The different peptides are: Val30 (black), Cys30 (brown), Thr30 (green), and Ser30 (cyan). The wavenumber shifts relative to the peptide with a valine at position 30 are indicated.
 C and
C and  O) labeled carbonyl group of N-Methylacetamide is depicted in red.
O) labeled carbonyl group of N-Methylacetamide is depicted in red.
 rotamers as a function of side-chain exposure in (Upper) helices and (Lower) nonhelical elements. The data set was a nonhomologous representation of all solved water-soluble proteins (37). Exposure was calculated as a ratio between the exposure of the specific serine and the maximum exposure.
rotamers as a function of side-chain exposure in (Upper) helices and (Lower) nonhelical elements. The data set was a nonhomologous representation of all solved water-soluble proteins (37). Exposure was calculated as a ratio between the exposure of the specific serine and the maximum exposure.References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

