Mapping posttranscriptional regulation of the human glycome uncovers microRNA defining the glycocode
- PMID: 24591635
- PMCID: PMC3964104
- DOI: 10.1073/pnas.1321524111
Mapping posttranscriptional regulation of the human glycome uncovers microRNA defining the glycocode
Abstract
Cell surface glycans form a critical interface with the biological milieu, informing diverse processes from the inflammatory cascade to cellular migration. Assembly of discrete carbohydrate structures requires the coordinated activity of a repertoire of proteins, including glycosyltransferases and glycosidases. Little is known about the regulatory networks controlling this complex biosynthetic process. Recent work points to a role for microRNA (miRNA) in the regulation of specific glycan biosynthetic enzymes. Herein we take a unique systems-based approach to identify connections between miRNA and the glycome. By using our glycomic analysis platform, lectin microarrays, we identify glycosylation signatures in the NCI-60 cell panel that point to the glycome as a direct output of genomic information flow. Integrating our glycomic dataset with miRNA data, we map miRNA regulators onto genes in glycan biosynthetic pathways (glycogenes) that generate the observed glycan structures. We validate three of these predicted miRNA/glycogene regulatory networks: high mannose, fucose, and terminal β-GalNAc, identifying miRNA regulation that would not have been observed by traditional bioinformatic methods. Overall, our work reveals critical nodes in the global glycosylation network accessible to miRNA regulation, providing a bridge between miRNA-mediated control of cell phenotype and the glycome.
Keywords: NCI-60; carbohydrate biosynthesis; epigenetics; glycan regulation; systems biology.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


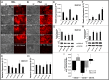


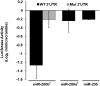
References
-
- Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem. 2010;79:351–379. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

