Regulatory network features in Listeria monocytogenes-changing the way we talk
- PMID: 24592357
- PMCID: PMC3924034
- DOI: 10.3389/fcimb.2014.00014
Regulatory network features in Listeria monocytogenes-changing the way we talk
Abstract
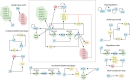
Our understanding of how pathogens shape their gene expression profiles in response to environmental changes is ever growing. Advances in Bioinformatics have made it possible to model complex systems and integrate data from variable sources into one large regulatory network. In these analyses, regulatory networks are typically broken down into regulatory motifs such as feed-forward loops (FFL) or auto-regulatory feedbacks, which serves to simplify the structure, while the functional implications of different regulatory motifs allow to make informed assumptions about the function of a specific regulatory pathway. Here we review the basic concepts of network features and use this language to break down the regulatory networks that govern the interactions between the main regulators of stress response, virulence, and transmission in Listeria monocytogenes. We point out the advantage that taking a "systems approach" could have for our understanding of gene functions, the detection of distant regulatory inputs, interspecies comparisons, and co-expression.
Keywords: Listeria monocytogenes; PrfA; SigB; network motif; regulatory network.
Figures

References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

