Interaction between transactivation domain of p53 and middle part of TBP-like protein (TLP) is involved in TLP-stimulated and p53-activated transcription from the p21 upstream promoter
- PMID: 24594805
- PMCID: PMC3940844
- DOI: 10.1371/journal.pone.0090190
Interaction between transactivation domain of p53 and middle part of TBP-like protein (TLP) is involved in TLP-stimulated and p53-activated transcription from the p21 upstream promoter
Abstract
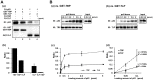
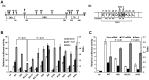
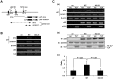
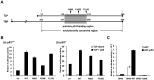
TBP-like protein (TLP) is involved in transcriptional activation of an upstream promoter of the human p21 gene. TLP binds to p53 and facilitates p53-activated transcription from the upstream promoter. In this study, we clarified that in vitro affinity between TLP and p53 is about one-third of that between TBP and p53. Extensive mutation analyses revealed that the TLP-stimulated function resides in transcription activating domain 1 (TAD1) in the N-terminus of p53. Among the mutants, #22.23, which has two amino acid substitutions in TAD1, exhibited a typical mutant phenotype. Moreover, #22.23 exhibited the strongest mutant phenotype for TLP-binding ability. It is thus thought that TLP-stimulated and p53-dependent transcriptional activation is involved in TAD1 binding of TLP. #22.23 had a decreased transcriptional activation function, especially for the upstream promoter of the endogenous p21 gene, compared with wild-type p53. This mutant did not facilitate p53-dependent growth repression and etoposide-mediated cell-death as wild-type p53 does. Moreover, mutation analysis revealed that middle part of TLP, which is requited for p53 binding, is involved in TLP-stimulated and p53-dependent promoter activation and cell growth repression. These results suggest that activation of the p21 upstream promoter is mediated by interaction between specific regions of TLP and p53.
Conflict of interest statement
Figures



Similar articles
-
Activity of the upstream TATA-less promoter of the p21(Waf1/Cip1) gene depends on transcription factor IIA (TFIIA) in addition to TFIIA-reactive TBP-like protein.FEBS J. 2014 Jul;281(14):3126-37. doi: 10.1111/febs.12848. Epub 2014 Jun 6. FEBS J. 2014. PMID: 24835508 Free PMC article.
-
TATA-binding protein (TBP)-like protein is required for p53-dependent transcriptional activation of upstream promoter of p21Waf1/Cip1 gene.J Biol Chem. 2012 Jun 8;287(24):19792-803. doi: 10.1074/jbc.M112.369629. Epub 2012 Apr 16. J Biol Chem. 2012. PMID: 22511763 Free PMC article.
-
Vertebrate TBP-like protein (TLP/TRF2/TLF) stimulates TATA-less terminal deoxynucleotidyl transferase promoters in a transient reporter assay, and TFIIA-binding capacity of TLP is required for this function.Nucleic Acids Res. 2003 Apr 15;31(8):2127-33. doi: 10.1093/nar/gkg315. Nucleic Acids Res. 2003. PMID: 12682363 Free PMC article.
-
TATA-binding Protein (TBP)-like Protein Is Engaged in Etoposide-induced Apoptosis through Transcriptional Activation of Human TAp63 Gene.J Biol Chem. 2009 Dec 18;284(51):35433-40. doi: 10.1074/jbc.M109.050047. J Biol Chem. 2009. PMID: 19858204 Free PMC article.
-
p53-dependent down-regulation of telomerase is mediated by p21waf1.J Biol Chem. 2004 Dec 3;279(49):50976-85. doi: 10.1074/jbc.M402502200. Epub 2004 Sep 15. J Biol Chem. 2004. PMID: 15371422
Cited by
-
Nucleosome distortion as a possible mechanism of transcription activation domain function.Epigenetics Chromatin. 2016 Sep 20;9:40. doi: 10.1186/s13072-016-0092-2. eCollection 2016. Epigenetics Chromatin. 2016. PMID: 27679670 Free PMC article. Review.
-
TBP-like Protein (TLP) Disrupts the p53-MDM2 Interaction and Induces Long-lasting p53 Activation.J Biol Chem. 2017 Feb 24;292(8):3201-3212. doi: 10.1074/jbc.M116.763318. Epub 2017 Jan 12. J Biol Chem. 2017. PMID: 28082682 Free PMC article.
-
TBP-like protein (TLP) interferes with Taspase1-mediated processing of TFIIA and represses TATA box gene expression.Nucleic Acids Res. 2015 Jul 27;43(13):6285-98. doi: 10.1093/nar/gkv576. Epub 2015 Jun 1. Nucleic Acids Res. 2015. PMID: 26038314 Free PMC article.
-
Activity of the upstream TATA-less promoter of the p21(Waf1/Cip1) gene depends on transcription factor IIA (TFIIA) in addition to TFIIA-reactive TBP-like protein.FEBS J. 2014 Jul;281(14):3126-37. doi: 10.1111/febs.12848. Epub 2014 Jun 6. FEBS J. 2014. PMID: 24835508 Free PMC article.
References
-
- el-Deiry WS, Tokino T, Velculescu VE, Levy DB, Parsons R, et al. (1993) WAF1, a potential mediator of p53 tumor suppression. Cell 75: 817–825. - PubMed
-
- Vousden KH (2002) Activation of the p53 tumor suppressor protein. Biochim Biophys Acta 14: 47–59. - PubMed
-
- Vousden KH, Lu X (2002) Live or let die: the cell's response to p53. Nat Rev Cancer 2: 594–604. - PubMed
-
- Hirao A, Kong YY, Matsuoka S, Wakeham A, Ruland J, et al. (2000) DNA Damage-Induced Activation of p53 by the Checkpoint Kinase Chk2. Science 287: 1824–1827. - PubMed
-
- Haupt Y, Maya R, Kazaz A, Oren M (1997) Mdm2 promotes the rapid degradation of p53. Nature 387: 296–299. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

