Osteosarcoma microenvironment: whole-slide imaging and optimized antigen detection overcome major limitations in immunohistochemical quantification
- PMID: 24594971
- PMCID: PMC3940945
- DOI: 10.1371/journal.pone.0090727
Osteosarcoma microenvironment: whole-slide imaging and optimized antigen detection overcome major limitations in immunohistochemical quantification
Abstract
Background: In osteosarcoma survival rates could not be improved over the last 30 years. Novel biomarkers are warranted to allow risk stratification of patients for more individual treatment following initial diagnosis. Although previous studies of the tumor microenvironment have identified promising candidates, novel biomarkers have not been translated into routine histopathology. Substantial difficulties regarding immunohistochemical detection and quantification of antigens in decalcified and heterogeneous osteosarcoma might largely explain this translational short-coming. Furthermore, we hypothesized that conventional hot spot analysis is often not representative for the whole section when applied to heterogeneous tissues like osteosarcoma. We aimed to overcome these difficulties for major biomarkers of the immunovascular microenvironment.
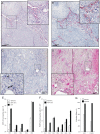
Methods: Immunohistochemistry was systematically optimized for cell surface (CD31, CD8) and intracellular antigens (FOXP3) including evaluation of 200 different antigen retrieval conditions. Distribution patterns of these antigens were analyzed in formalin-fixed and paraffin-embedded samples from 120 high-grade central osteosarcoma biopsies and computer-assisted whole-slide analysis was compared with conventional quantification methods including hot spot analysis.
Results: More than 96% of osteosarcoma samples were positive for all antigens after optimization of immunohistochemistry. In contrast, standard immunohistochemistry retrieved false negative results in 35-65% of decalcified osteosarcoma specimens. Standard hot spot analysis was applicable for homogeneous distributed FOXP3+ and CD8+ cells. However, heterogeneous distribution of vascular CD31 did not allow reliable quantification with hot spot analysis in 85% of all samples. Computer-assisted whole-slide analysis of total CD31- immunoreactive area proved as the most appropriate quantification method.
Conclusion: Standard staining and quantification procedures are not applicable in decalcified formalin-fixed and paraffin-embedded samples for major parameters of the immunovascular microenvironment in osteosarcoma. Whole-slide imaging and optimized antigen retrieval overcome these limitations.
Conflict of interest statement
Figures



References
-
- Clark JC, Dass CR, Choong PF (2008) A review of clinical and molecular prognostic factors in osteosarcoma. J Cancer Res Clin Oncol 134: 281–297. - PubMed
-
- Klein MJ, Siegal GP (2006) Osteosarcoma: anatomic and histologic variants. Am J Clin Pathol 125: 555–581. - PubMed
-
- Buckanovich RJ, Facciabene A, Kim S, Benencia F, Sasaroli D, et al. (2008) Endothelin B receptor mediates the endothelial barrier to T cell homing to tumors and disables immune therapy. Nat Med 14: 28–36. - PubMed
-
- Curiel TJ, Cheng P, Mottram P, Alvarez X, Moons L, et al. (2004) Dendritic cell subsets differentially regulate angiogenesis in human ovarian cancer. Cancer Res 64: 5535–5538. - PubMed
-
- Facciabene A, Peng X, Hagemann IS, Balint K, Barchetti A, et al. (2011) Tumour hypoxia promotes tolerance and angiogenesis via CCL28 and T(reg) cells. Nature 475: 226–230. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

