Three-dimensional structural aspects of protein-polysaccharide interactions
- PMID: 24595239
- PMCID: PMC3975366
- DOI: 10.3390/ijms15033768
Three-dimensional structural aspects of protein-polysaccharide interactions
Abstract
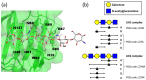
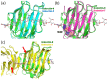
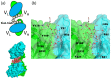
Linear polysaccharides are typically composed of repeating mono- or disaccharide units and are ubiquitous among living organisms. Polysaccharide diversity arises from chain-length variation, branching, and additional modifications. Structural diversity is associated with various physiological functions, which are often regulated by cognate polysaccharide-binding proteins. Proteins that interact with linear polysaccharides have been identified or developed, such as galectins and polysaccharide-specific antibodies, respectively. Currently, data is accumulating on the three-dimensional structure of polysaccharide-binding proteins. These proteins are classified into two types: exo-type and endo-type. The former group specifically interacts with the terminal units of polysaccharides, whereas the latter with internal units. In this review, we describe the structural aspects of exo-type and endo-type protein-polysaccharide interactions. Further, we discuss the structural basis for affinity and specificity enhancement in the face of inherently weak binding interactions.
Figures



References
-
- Lee R.T., Lee Y.C. Affinity enhancement by multivalent lectin-carbohydrate interaction. Glycoconj. J. 2000;17:543–551. - PubMed
-
- Fred Brewer C. Binding and cross-linking properties of galectins. Biochim. Biophys. Acta. 2002;1572:255–262. - PubMed
-
- Bourne Y., Bolgiano B., Liao D.I., Strecker G., Cantau P., Herzberg O., Feizi T., Cambillau C. Crosslinking of mammalian lectin (galectin-1) by complex biantennary saccharides. Nat. Struct. Biol. 1994;1:863–870. - PubMed
-
- Gupta D., Bhattacharyya L., Fant J., Macaluso F., Sabesan S., Brewer C.F. Observation of unique cross-linked lattices between multiantennary carbohydrates and soybean lectin Presence of pseudo-2-fold axes of symmetry in complex type carbohydrates. Biochemistry. 1994;33:7495– 7504. - PubMed
-
- Olsen L.R., Dessen A., Gupta D., Sabesan S., Sacchettini J.C., Brewer C.F. X-ray crystallographic studies of unique cross-linked lattices between four isomeric biantennary oligosaccharides and soybean agglutinin. Biochemistry. 1997;36:15073–15080. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

