ATP requirement for chloroplast protein import is set by the Km for ATP hydrolysis of stromal Hsp70 in Physcomitrella patens
- PMID: 24596240
- PMCID: PMC4001381
- DOI: 10.1105/tpc.113.121822
ATP requirement for chloroplast protein import is set by the Km for ATP hydrolysis of stromal Hsp70 in Physcomitrella patens
Abstract
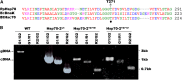
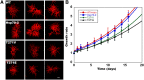
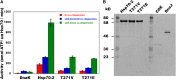
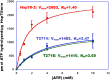
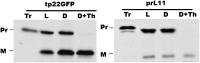
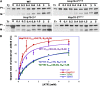
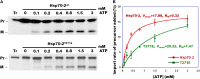
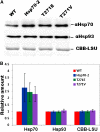
The 70-kD family of heat shock proteins (Hsp70s) is involved in a number of seemingly disparate cellular functions, including folding of nascent proteins, breakup of misfolded protein aggregates, and translocation of proteins across membranes. They act through the binding and release of substrate proteins, accompanied by hydrolysis of ATP. Chloroplast stromal Hsp70 plays a crucial role in the import of proteins into plastids. Mutations of an ATP binding domain Thr were previously reported to result in an increase in the Km for ATP and a decrease in the enzyme's kcat. To ask which chloroplast stromal chaperone, Hsp70 or Hsp93, both of which are ATPases, dominates the energetics of the motor responsible for protein import, we made transgenic moss (Physcomitrella patens) harboring the Km-altering mutation in the essential stromal Hsp70-2 and measured the effect on the amount of ATP required for protein import into chloroplasts. Here, we report that increasing the Km for ATP hydrolysis of Hsp70 translated into an increased Km for ATP usage by chloroplasts for protein import. This thus directly demonstrates that the ATP-derived energy long known to be required for chloroplast protein import is delivered via the Hsp70 chaperones and that the chaperone's ATPase activity dominates the energetics of the reaction.
Figures
References
-
- Alder N.N., Theg S.M. (2003). Protein transport via the cpTat pathway displays cooperativity and is stimulated by transport-incompetent substrate. FEBS Lett. 540: 96–100. - PubMed
-
- Aronsson H., Jarvis P. (2002). A simple method for isolating import-competent Arabidopsis chloroplasts. FEBS Lett. 529: 215–220. - PubMed
-
- Bagola K., Mehnert M., Jarosch E., Sommer T. (2011). Protein dislocation from the ER. Biochim. Biophys. Acta 1808: 925–936. - PubMed
-
- Becker T., Böttinger L., Pfanner N. (2012). Mitochondrial protein import: From transport pathways to an integrated network. Trends Biochem. Sci. 37: 85–91. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

