Molecular functions of the TLE tetramerization domain in Wnt target gene repression
- PMID: 24596249
- PMCID: PMC4000089
- DOI: 10.1002/embj.201387188
Molecular functions of the TLE tetramerization domain in Wnt target gene repression
Abstract
Wnt signaling activates target genes by promoting association of the co-activator β-catenin with TCF/LEF transcription factors. In the absence of β-catenin, target genes are silenced by TCF-mediated recruitment of TLE/Groucho proteins, but the molecular basis for TLE/TCF-dependent repression is unclear. We describe the unusual three-dimensional structure of the N-terminal Q domain of TLE1 that mediates tetramerization and binds to TCFs. We find that differences in repression potential of TCF/LEFs correlates with their affinities for TLE-Q, rather than direct competition between β-catenin and TLE for TCFs as part of an activation-repression switch. Structure-based mutation of the TLE tetramer interface shows that dimers cannot mediate repression, even though they bind to TCFs with the same affinity as tetramers. Furthermore, the TLE Q tetramer, not the dimer, binds to chromatin, specifically to K20 methylated histone H4 tails, suggesting that the TCF/TLE tetramer complex promotes structural transitions of chromatin to mediate repression.
Figures

The primary structure of human TLE1 showing domain boundaries.
The structure of the TLE1 tetramerization domain comprising residues 23–136 in chains A (light blue) and B (red), and 23–133 in chains C (orange) and D (green) are shown; the remaining residues are disordered. Hydrophobic side chains are shown in stick representation, and the N- and C-termini marked.
Hydrophobic interactions between chain C (orange) and chain D (green) involving the α2 and α3 helices. The 120° orientation between α2 and α3 is fixed by a salt bridge between E118 of one chain and R84 of its partner.
Glutamine residues (shown in stick representation) are mostly clustered on helices α2 and α3.
Close-up of the TLE1-Q tetramer interface.
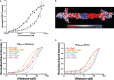
SAXS analysis of TLE120–156. The solid black line shows the experimentally determined scattering curve of TLE120–156 at 5 mg/ml. The Rg obtained from the Guinier plot = 70 Å, and Dmax = 227 Å as calculated with Autognom (Semenyuk & Svergun, 1991). These values match closely the Rg = 69 Å and Dmax = 217 Å calculated from the extended tetramer model (green line; χ2 = 3.1) versus Rg = 35 Å and Dmax = 162 Å for the compact tetramer (red line; χ2 = 6.9; see Supplementary Fig S1B).
Gel filtration of the TLE120–156 tetramer and L26D, I29D dimer mutant. Purified proteins were run on a Superdex S200 gel filtration column. Due to their large hydrodynamic radius (axial ratio ˜10:1), the proteins run at apparent molecular weights of 173 kDa, and 53 kDa.
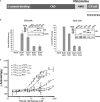
Primary structure of the TCF-LEF family of proteins, showing the N-terminal β-catenin binding domain, the context-dependent regulatory domain (CRD), and the HMG box that binds DNA. TCF3 and TCF4 also have extensions that arise from alternative splicing. Residue numbers for domain boundaries are shown for the XTCF3 protein used in this study. The equivalent sequence numbers of other family members can be found in Supplementary Fig S2.
Luciferase reporter assay in HEK293 cells (left) and Cos1 cells (right) shows differential abilities of transfected HA-tagged TCF/LEFs to activate the Wnt reporter SuperTOPFlash when co-transfected with β-catenin. The graphs for each cell line are representative of three replicates. The Western blots (anti-HA) shown in the insets reveal the relative expression levels of the transfected TCF/LEF constructs and show that the activity differences are not related to protein levels. The band marked * is a breakdown product of TCF3. Error bars represent the standard deviation of triplicate measurements.
Fluorescence anisotropy analysis of TLE120–156 binding to different TCF/LEFs. Increasing amounts of TCF/LEF ligands were titrated against labeled TLE120–156. The change in anisotropy was measured and plotted as a function of concentration. Error bars represent standard deviation of triplicate measurements.
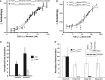
Binding of TLE120–156 tetramer to the TCF3(1–330): β-catenin complex or TCF31–330 measured by fluorescence anisotropy. Error bars represent standard deviation of n = 3.
Binding of TLE120–156 dimer mutant to and TCF31–330 and TCF41–330. Error bars represent standard deviation of n = 3.
Overexpression of TCF3 leads to significant repression of reporter activity in HEK293T cells. Cells were treated with 50 ng of purified Wnt3a 24 h after co-transfection with or without TCF3. Error bars represent standard deviation of n = 3.
TLE tetramer but not the dimer can repress reporter activity in HEK293T cells. Cells were treated with 100 ng of purified Wnt3a 24 h after co-transfection with β-catenin, TCF3 and indicated amounts of DNA constructs encoding either TLE1 dimer or TLE1 tetramer. Error bars represent standard deviation of n = 3. Inset Western blot shows levels of transfected TLE1 tetramer or dimer. Immunofluorescence demonstrating nuclear localization of transfectred TLE1 constructs is shown in Supplementary Fig S3.

References
-
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. - PMC - PubMed
-
- Barbera AJ, Chodaparambil JV, Kelley-Clarke B, Joukov V, Walter JC, Luger K, Kaye KM. The nucleosomal surface as a docking station for Kaposi's sarcoma herpesvirus LANA. Science. 2006;311:856–861. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

