Architecture and function of metallopeptidase catalytic domains
- PMID: 24596965
- PMCID: PMC3926739
- DOI: 10.1002/pro.2400
Architecture and function of metallopeptidase catalytic domains
Abstract
The cleavage of peptide bonds by metallopeptidases (MPs) is essential for life. These ubiquitous enzymes participate in all major physiological processes, and so their deregulation leads to diseases ranging from cancer and metastasis, inflammation, and microbial infection to neurological insults and cardiovascular disorders. MPs cleave their substrates without a covalent intermediate in a single-step reaction involving a solvent molecule, a general base/acid, and a mono- or dinuclear catalytic metal site. Most monometallic MPs comprise a short metal-binding motif (HEXXH), which includes two metal-binding histidines and a general base/acid glutamate, and they are grouped into the zincin tribe of MPs. The latter divides mainly into the gluzincin and metzincin clans. Metzincins consist of globular ∼ 130-270-residue catalytic domains, which are usually preceded by N-terminal pro-segments, typically required for folding and latency maintenance. The catalytic domains are often followed by C-terminal domains for substrate recognition and other protein-protein interactions, anchoring to membranes, oligomerization, and compartmentalization. Metzincin catalytic domains consist of a structurally conserved N-terminal subdomain spanning a five-stranded β-sheet, a backing helix, and an active-site helix. The latter contains most of the metal-binding motif, which is here characteristically extended to HEXXHXXGXX(H,D). Downstream C-terminal subdomains are generally shorter, differ more among metzincins, and mainly share a conserved loop--the Met-turn--and a C-terminal helix. The accumulated structural data from more than 300 deposited structures of the 12 currently characterized metzincin families reviewed here provide detailed knowledge of the molecular features of their catalytic domains, help in our understanding of their working mechanisms, and form the basis for the design of novel drugs.
Figures



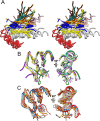
References
-
- Barrett AJ, Rawlings ND, Salvesen G, Woessner JF. In: Handbook of proteolytic enzymes. Rawlings ND, Salvesen G, editors. Oxford (UK): Academic Press; 2013. pp. Ii–Iiv.
-
- Neurath H. Limited proteolysis and zymogen activation. In: Reich E, Rifkins DB, Shaw E, editors. Proteases and biological control. New York: Cold Spring Harbor Laboratory Press; 1975. pp. 51–64.
-
- López-Otín C, Overall CM. Protease degradomics: a new challenge for proteomics. Nat Rev Mol Cell Biol. 2002;3:509–519. - PubMed
-
- Kheradmand F, Werb Z. Shedding light on sheddases: role in growth and development. BioEssays. 2002;24:8–12. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

