Recognition of floral homeotic MADS domain transcription factors by a phytoplasmal effector, phyllogen, induces phyllody
- PMID: 24597566
- PMCID: PMC4282529
- DOI: 10.1111/tpj.12495
Recognition of floral homeotic MADS domain transcription factors by a phytoplasmal effector, phyllogen, induces phyllody
Abstract
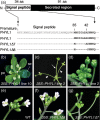
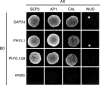
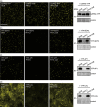
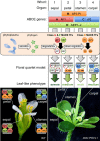
Plant pathogens alter the course of plant developmental processes, resulting in abnormal morphology in infected host plants. Phytoplasmas are unique plant-pathogenic bacteria that transform plant floral organs into leaf-like structures and cause the emergence of secondary flowers. These distinctive symptoms have attracted considerable interest for many years. Here, we revealed the molecular mechanisms of the floral symptoms by focusing on a phytoplasma-secreted protein, PHYL1, which induces morphological changes in flowers that are similar to those seen in phytoplasma-infected plants. PHYL1 is a homolog of the phytoplasmal effector SAP54 that also alters floral development. Using yeast two-hybrid and in planta transient co-expression assays, we found that PHYL1 interacts with and degrades the floral homeotic MADS domain proteins SEPALLATA3 (SEP3), APETALA1 (AP1) and CAULIFLOWER (CAL). This degradation of MADS domain proteins was dependent on the ubiquitin-proteasome pathway. The expression of floral development genes downstream of SEP3 and AP1 was disrupted in 35S::PHYL1 transgenic plants. PHYL1 was genetically and functionally conserved among other phytoplasma strains and species. We designate PHYL1, SAP54 and their homologs as members of the phyllody-inducing gene family of 'phyllogens'.
Keywords: Arabidopsis; MADS domain proteins; floral development; floral quartet model; phyllody; phytoplasma.
© 2014 The Authors.The Plant Journal published by Society for Experimental Biology and John Wiley & Sons Ltd.
Figures



References
-
- Bertaccini A, Duduk B. Phytoplasma and phytoplasma diseases: a review of recent research. Phytopathol. Mediterr. 2009;48:355–378.
-
- Bowman JL, Alvarez J, Weigel D, Meyerowitz EM, Smyth DR. Control of flower development in Arabidopsis thaliana by APETALA1 and interacting genes. Development. 1993;119:721–743.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

