Post-transcriptional regulatory network of epithelial-to-mesenchymal and mesenchymal-to-epithelial transitions
- PMID: 24598126
- PMCID: PMC3973872
- DOI: 10.1186/1756-8722-7-19
Post-transcriptional regulatory network of epithelial-to-mesenchymal and mesenchymal-to-epithelial transitions
Abstract
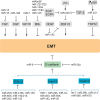
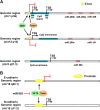
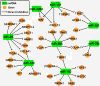
Epithelial-to-mesenchymal transition (EMT) and its reverse process, mesenchymal-to-epithelial transition (MET), play important roles in embryogenesis, stem cell biology, and cancer progression. EMT can be regulated by many signaling pathways and regulatory transcriptional networks. Furthermore, post-transcriptional regulatory networks regulate EMT; these networks include the long non-coding RNA (lncRNA) and microRNA (miRNA) families. Specifically, the miR-200 family, miR-101, miR-506, and several lncRNAs have been found to regulate EMT. Recent studies have illustrated that several lncRNAs are overexpressed in various cancers and that they can promote tumor metastasis by inducing EMT. MiRNA controls EMT by regulating EMT transcription factors or other EMT regulators, suggesting that lncRNAs and miRNA are novel therapeutic targets for the treatment of cancer. Further efforts have shown that non-coding-mediated EMT regulation is closely associated with epigenetic regulation through promoter methylation (e.g., miR-200 or miR-506) and protein regulation (e.g., SET8 via miR-502). The formation of gene fusions has also been found to promote EMT in prostate cancer. In this review, we discuss the post-transcriptional regulatory network that is involved in EMT and MET and how targeting EMT and MET may provide effective therapeutics for human disease.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

