Bacterial promoter repression by DNA looping without protein-protein binding competition
- PMID: 24598256
- PMCID: PMC4027209
- DOI: 10.1093/nar/gku180
Bacterial promoter repression by DNA looping without protein-protein binding competition
Abstract
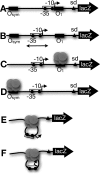
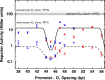
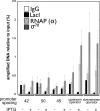
The Escherichia coli lactose operon provides a paradigm for understanding gene control by DNA looping where the lac repressor (LacI) protein competes with RNA polymerase for DNA binding. Not all promoter loops involve direct competition between repressor and RNA polymerase. This raises the possibility that positioning a promoter within a tightly constrained DNA loop is repressive per se, an idea that has previously only been considered in vitro. Here, we engineer living E. coli bacteria to measure repression due to promoter positioning within such a tightly constrained DNA loop in the absence of protein-protein binding competition. We show that promoters held within such DNA loops are repressed ∼100-fold, with up to an additional ∼10-fold repression (∼1000-fold total) dependent on topological positioning of the promoter on the inner or outer face of the DNA loop. Chromatin immunoprecipitation data suggest that repression involves inhibition of both RNA polymerase initiation and elongation. These in vivo results show that gene repression can result from tightly looping promoter DNA even in the absence of direct competition between repressor and RNA polymerase binding.
© The Author(s) 2014. Published by Oxford University Press.
Figures
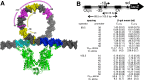

References
-
- Müller-Hill B. The lac Operon: a Short History of a Genetic Paradigm. Berlin: Walter de Gruyter; 1996.
-
- Jacob F., Monod J. Genetic regulatory mechanisms in the synthesis of proteins. J. Mol. Biol. 1961;3:318–356. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

