Community-wide transcriptome of the oral microbiome in subjects with and without periodontitis
- PMID: 24599074
- PMCID: PMC4817619
- DOI: 10.1038/ismej.2014.23
Community-wide transcriptome of the oral microbiome in subjects with and without periodontitis
Abstract
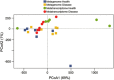
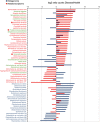
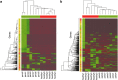
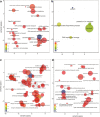
Despite increasing knowledge on phylogenetic composition of the human microbiome, our understanding of the in situ activities of the organisms in the community and their interactions with each other and with the environment remains limited. Characterizing gene expression profiles of the human microbiome is essential for linking the role of different members of the bacterial communities in health and disease. The oral microbiome is one of the most complex microbial communities in the human body and under certain circumstances, not completely understood, the healthy microbial community undergoes a transformation toward a pathogenic state that gives rise to periodontitis, a polymicrobial inflammatory disease. We report here the in situ genome-wide transcriptome of the subgingival microbiome in six periodontally healthy individuals and seven individuals with periodontitis. The overall picture of metabolic activities showed that iron acquisition, lipopolysaccharide synthesis and flagellar synthesis were major activities defining disease. Unexpectedly, the vast majority of virulence factors upregulated in subjects with periodontitis came from organisms that are not considered major periodontal pathogens. One of the organisms whose gene expression profile was characterized was the uncultured candidate division TM7, showing an upregulation of putative virulence factors in the diseased community. These data enhance understanding of the core activities that are characteristic of periodontal disease as well as the role that individual organisms in the subgingival community play in periodontitis.
Figures
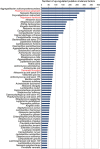
References
-
- Bender MH, Weiser JN. (2006). The atypical amino-terminal LPNTG-containing domain of the pneumococcal human IgA1-specific protease is required for proper enzyme localization and function. Mol Microbiol 61: 526–543. - PubMed
-
- Booijink CCGM, Boekhorst J, Zoetendal EG, Smidt H, Kleerebezem M, de Vos WM. (2010). Metatranscriptome analysis of the human fecal microbiota reveals subject-specific expression profiles, with genes encoding proteins involved in carbohydrate metabolism being dominantly expressed. Appl Environ Microbiol 76: 5533–5540. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

