Plants utilize a highly conserved system for repair of NADH and NADPH hydrates
- PMID: 24599492
- PMCID: PMC4012604
- DOI: 10.1104/pp.114.236539
Plants utilize a highly conserved system for repair of NADH and NADPH hydrates
Abstract
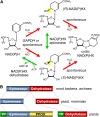
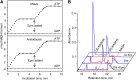
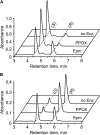
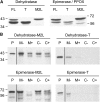
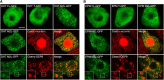
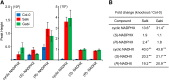
NADH and NADPH undergo spontaneous and enzymatic reactions that produce R and S forms of NAD(P)H hydrates [NAD(P)HX], which are not electron donors and inhibit various dehydrogenases. In bacteria, yeast (Saccharomyces cerevisiae), and mammals, these hydrates are repaired by the tandem action of an ADP- or ATP-dependent dehydratase that converts (S)-NAD(P)HX to NAD(P)H and an epimerase that facilitates interconversion of the R and S forms. Plants have homologs of both enzymes, the epimerase homolog being fused to the vitamin B6 salvage enzyme pyridoxine 5'-phosphate oxidase. Recombinant maize (Zea mays) and Arabidopsis (Arabidopsis thaliana) NAD(P)HX dehydratases (GRMZM5G840928, At5g19150) were able to reconvert (S)-NAD(P)HX to NAD(P)H in an ATP-dependent manner. Recombinant maize and Arabidopsis epimerases (GRMZM2G061988, At5g49970) rapidly interconverted (R)- and (S)-NAD(P)HX, as did a truncated form of the Arabidopsis epimerase lacking the pyridoxine 5'-phosphate oxidase domain. All plant NAD(P)HX dehydratase and epimerase sequences examined had predicted organellar targeting peptides with a potential second start codon whose use would eliminate the targeting peptide. In vitro transcription/translation assays confirmed that both start sites were used. Dual import assays with purified pea (Pisum sativum) chloroplasts and mitochondria, and subcellular localization of GFP fusion constructs in tobacco (Nicotiana tabacum) suspension cells, indicated mitochondrial, plastidial, and cytosolic localization of the Arabidopsis epimerase and dehydratase. Ablation of the Arabidopsis dehydratase gene raised seedling levels of all NADHX forms by 20- to 40-fold, and levels of one NADPHX form by 10- to 30-fold. We conclude that plants have a canonical two-enzyme NAD(P)HX repair system that is directed to three subcellular compartments via the use of alternative translation start sites.
Figures
References
-
- Acheson SA, Kirkman HN, Wolfenden R. (1988) Equilibrium of 5,6-hydration of NADH and mechanism of ATP-dependent dehydration. Biochemistry 27: 7371–7375 - PubMed
-
- Bradford MM. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 - PubMed
-
- Brandizzi F, Irons S, Kearns A, Hawes C. (2003) BY-2 cells: culture and transformation for live cell imaging. Curr Protoc Cell Biol Chapter 1: 1.7.1–1.7.16 - PubMed
-
- Chabregas SM, Luche DD, Van Sluys MA, Menck CF, Silva-Filho MC. (2003) Differential usage of two in-frame translational start codons regulates subcellular localization of Arabidopsis thaliana THI1. J Cell Sci 116: 285–291 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

