Endothelial Kruppel-like factor 4 regulates angiogenesis and the Notch signaling pathway
- PMID: 24599951
- PMCID: PMC4002108
- DOI: 10.1074/jbc.M113.530956
Endothelial Kruppel-like factor 4 regulates angiogenesis and the Notch signaling pathway
Abstract
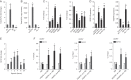
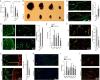
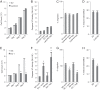
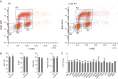
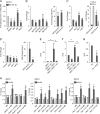
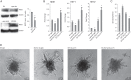
Regulation of endothelial cell biology by the Notch signaling pathway (Notch) is essential to vascular development, homeostasis, and sprouting angiogenesis. Although Notch determines cell fate and differentiation in a wide variety of cells, the molecular basis of upstream regulation of Notch remains poorly understood. Our group and others have implicated the Krüppel-like factor family of transcription factors as critical regulators of endothelial function. Here, we show that Krüppel-like factor 4 (KLF4) is a central regulator of sprouting angiogenesis via regulating Notch. Using a murine model in which KLF4 is overexpressed exclusively in the endothelium, we found that sustained expression of KLF4 promotes ineffective angiogenesis leading to diminished tumor growth independent of endothelial cell proliferation or cell cycling effects. These tumors feature increased vessel density yet are hypoperfused, leading to tumor hypoxia. Mechanistically, we show that KLF4 differentially regulates expression of Notch receptors, ligands, and target genes. We also demonstrate that KLF4 limits cleavage-mediated activation of Notch1. Finally, we rescue Notch target gene expression and the KLF4 sprouting angiogenesis phenotype by supplementation of DLL4 recombinant protein. Identification of this hitherto undiscovered role of KLF4 implicates this transcription factor as a critical regulator of Notch, tumor angiogenesis, and sprouting angiogenesis.
Keywords: ATPases; Actin; Arabidopsis; Kinetics; Molecular Motors; Myosin.
Figures


References
-
- Iso T., Hamamori Y., Kedes L. (2003) Notch signaling in vascular development. Arterioscler. Thromb. Vasc. Biol. 23, 543–553 - PubMed
-
- Hrabĕ de Angelis M., McIntyre J., 2nd, Gossler A. (1997) Maintenance of somite borders in mice requires the Delta homologue DLL1. Nature 386, 717–721 - PubMed
-
- Roca C., Adams R. H. (2007) Regulation of vascular morphogenesis by Notch signaling. Genes Dev. 21, 2511–2524 - PubMed
-
- Xue Y., Gao X., Lindsell C. E., Norton C. R., Chang B., Hicks C., Gendron-Maguire M., Rand E. B., Weinmaster G., Gridley T. (1999) Embryonic lethality and vascular defects in mice lacking the Notch ligand Jagged1. Hum. Mol. Genet. 8, 723–730 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

