Phosphatidic acid (PA)-preferring phospholipase A1 regulates mitochondrial dynamics
- PMID: 24599962
- PMCID: PMC4036285
- DOI: 10.1074/jbc.M113.531921
Phosphatidic acid (PA)-preferring phospholipase A1 regulates mitochondrial dynamics
Abstract
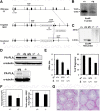
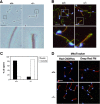
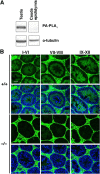
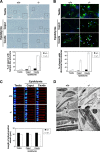
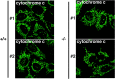
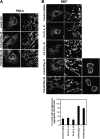
Recent studies have suggested that phosphatidic acid (PA), a cone-shaped phospholipid that can generate negative curvature of lipid membranes, participates in mitochondrial fusion. However, precise mechanisms underling the production and consumption of PA on the mitochondrial surface are not fully understood. Phosphatidic acid-preferring phospholipase A1 (PA-PLA1)/DDHD1 is the first identified intracellular phospholipase A1 and preferentially hydrolyzes PA in vitro. Its cellular and physiological functions have not been elucidated. In this study, we show that PA-PLA1 regulates mitochondrial dynamics. PA-PLA1, when ectopically expressed in HeLa cells, induced mitochondrial fragmentation, whereas its depletion caused mitochondrial elongation. The effects of PA-PLA1 on mitochondrial morphology appear to counteract those of MitoPLD, a mitochondrion-localized phospholipase D that produces PA from cardiolipin. Consistent with high levels of expression of PA-PLA1 in testis, PA-PLA1 knock-out mice have a defect in sperm formation. In PA-PLA1-deficient sperm, the mitochondrial structure is disorganized, and an abnormal gap structure exists between the middle and principal pieces. A flagellum is bent at that position, leading to a loss of motility. Our results suggest a possible mechanism of PA regulation of the mitochondrial membrane and demonstrate an in vivo function of PA-PLA1 in the organization of mitochondria during spermiogenesis.
Keywords: Membrane; Mitochondria; Phosphatidic Acid; Phospholipase A; Sperm.
Figures
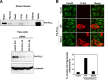
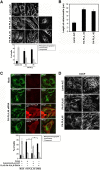
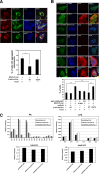
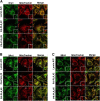
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

