Nuclear-cytoplasmic partitioning of cucumber mosaic virus protein 2b determines the balance between its roles as a virulence determinant and an RNA-silencing suppressor
- PMID: 24599997
- PMCID: PMC4019134
- DOI: 10.1128/JVI.00284-14
Nuclear-cytoplasmic partitioning of cucumber mosaic virus protein 2b determines the balance between its roles as a virulence determinant and an RNA-silencing suppressor
Abstract
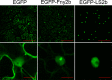
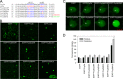
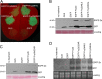
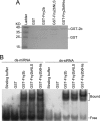
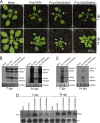
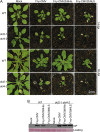
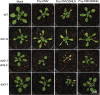
The Cucumber Mosaic Virus (CMV) 2b protein is an RNA-silencing suppressor that plays roles in CMV accumulation and virulence. The 2b proteins of subgroup IA CMV strains partition between the nucleus and cytoplasm, but the biological significance of this is uncertain. We fused an additional nuclear localization signal (NLS) to the 2b protein of subgroup IA strain Fny-CMV to create 2b-NLS and tested its effects on subcellular distribution, silencing, and virulence. The additional NLS enhanced 2b protein nuclear and nucleolar accumulation, but nuclear and nucleolar enrichment correlated with markedly diminished silencing suppressor activity in patch assays and abolished 2b protein-mediated disruption of microRNA activity in transgenic Arabidopsis. Nucleus/nucleolus-localized 2b protein possesses at least some ability to inhibit antiviral silencing, but this was not sufficient to prevent recovery from disease in younger, developing leaves in Arabidopsis. However, enhanced nuclear and nucleolar accumulation of 2b increased virulence and accelerated symptom appearance in older leaves. Experiments with Arabidopsis lines carrying mutant Dicer-like alleles demonstrated that compromised suppressor activity explained the diminished ability of 2b-NLS to enhance virus accumulation. Remarkably, the increased virulence that 2b-NLS engendered was unrelated to effects on microRNA- or short interfering RNA-regulated host functions. Thus, although nucleus- and nucleolus-localized 2b protein is less efficient at silencing suppression than cytoplasm-localized 2b, it enhances CMV virulence. We propose that partitioning of the 2b protein between the cytoplasmic and nuclear/nucleolar compartments allows CMV to regulate the balance between virus accumulation and damage to the host, presumably to maximize the benefit for the virus.
Importance: In this work, the main finding is that nucleus/nucleolus-localized 2b protein is strongly associated with CMV virulence, which is independent of its effect on small RNA pathways. Moreover, this work supports the contention that the silencing suppressor activity of CMV 2b protein is predominantly exerted by that portion of the 2b protein residing in the cytoplasm. Thus, we propose that partitioning of the 2b protein between the cytoplasmic and nuclear/nucleolar compartments allows CMV to regulate the balance between virus accumulation and damage to the host, presumably to maximize the benefit for the virus.
Figures

Similar articles
-
Strain-specific differences in the interactions of the cucumber mosaic virus 2b protein with the viral 1a and host Argonaute 1 proteins.J Virol. 2024 Sep 17;98(9):e0099324. doi: 10.1128/jvi.00993-24. Epub 2024 Aug 20. J Virol. 2024. PMID: 39162432 Free PMC article.
-
Self-interaction of the cucumber mosaic virus 2b protein plays a vital role in the suppression of RNA silencing and the induction of viral symptoms.Mol Plant Pathol. 2013 Oct;14(8):803-12. doi: 10.1111/mpp.12051. Epub 2013 Jun 19. Mol Plant Pathol. 2013. PMID: 23782515 Free PMC article.
-
Virus-induced necrosis is a consequence of direct protein-protein interaction between a viral RNA-silencing suppressor and a host catalase.Plant Physiol. 2011 Aug;156(4):2026-36. doi: 10.1104/pp.111.180042. Epub 2011 May 27. Plant Physiol. 2011. PMID: 21622812 Free PMC article.
-
Cucumber mosaic virus.Adv Virus Res. 2012;84:439-504. doi: 10.1016/B978-0-12-394314-9.00013-0. Adv Virus Res. 2012. PMID: 22682176 Review.
-
Nuclear RNA-binding proteins meet cytoplasmic viruses.RNA. 2025 Feb 19;31(3):444-451. doi: 10.1261/rna.080313.124. RNA. 2025. PMID: 39805659 Free PMC article. Review.
Cited by
-
Identification of the Potential Virulence Factors and RNA Silencing Suppressors of Mulberry Mosaic Dwarf-Associated Geminivirus.Viruses. 2018 Sep 3;10(9):472. doi: 10.3390/v10090472. Viruses. 2018. PMID: 30177616 Free PMC article.
-
CMV2b-AGO Interaction Is Required for the Suppression of RDR-Dependent Antiviral Silencing in Arabidopsis.Front Microbiol. 2016 Aug 24;7:1329. doi: 10.3389/fmicb.2016.01329. eCollection 2016. Front Microbiol. 2016. PMID: 27605926 Free PMC article.
-
Pepper CabZIP63 acts as a positive regulator during Ralstonia solanacearum or high temperature-high humidity challenge in a positive feedback loop with CaWRKY40.J Exp Bot. 2016 Apr;67(8):2439-51. doi: 10.1093/jxb/erw069. Epub 2016 Mar 1. J Exp Bot. 2016. PMID: 26936828 Free PMC article.
-
Plant viral proteins and fibrillarin: the link to complete the infective cycle.Mol Biol Rep. 2021 May;48(5):4677-4686. doi: 10.1007/s11033-021-06401-1. Epub 2021 May 25. Mol Biol Rep. 2021. PMID: 34036480 Review.
-
P25 and P37 proteins encoded by firespike leafroll-associated virus are viral suppressors of RNA silencing.Front Microbiol. 2022 Aug 16;13:964156. doi: 10.3389/fmicb.2022.964156. eCollection 2022. Front Microbiol. 2022. PMID: 36051767 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

